3SDM
 
 | | Structure of oligomeric kinase/RNase Ire1 in complex with an oligonucleotide | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1.
Bmc Biol., 9, 2011
|
|
3SDJ
 
 | | Structure of RNase-inactive point mutant of oligomeric kinase/RNase Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural and functional basis for RNA cleavage by Ire1.
Bmc Biol., 9, 2011
|
|
2M1R
 
 | | PHD domain of ING4 N214D mutant | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Blanco, F.J. | | Deposit date: | 2012-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional impact of cancer-associated mutations in the tumor suppressor protein ING4.
Carcinogenesis, 31, 2010
|
|
5TTV
 
 | | Jak3 with covalent inhibitor 6 | | Descriptor: | N-[3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl]propanamide, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
1SRX
 
 | |
5TTU
 
 | | Jak3 with covalent inhibitor 7 | | Descriptor: | 1-[(3aR,7aR)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydro-6H-pyrrolo[2,3-c]pyridin-6-yl]propan-1-one, SULFATE ION, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
5TTS
 
 | | Jak3 with covalent inhibitor 4 | | Descriptor: | 1-{(3R)-3-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}propan-1-one, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
7D7X
 
 | |
7D7W
 
 | |
7D82
 
 | |
7D81
 
 | |
7D7Y
 
 | |
7CXA
 
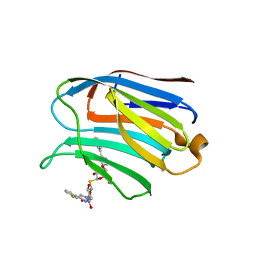 | | Structure of human Galectin-3 CRD in complex with TD-139 belonging to P31 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXC
 
 | |
7CXB
 
 | | Structure of mouse Galectin-3 CRD in complex with TD-139 belonging to P6522 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
8XZK
 
 | | Crystal structure of folE riboswitch | | Descriptor: | RNA (53-MER) | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZR
 
 | | Crystal structure of folE riboswitch with 8-NH2 Guanine | | Descriptor: | 8-AMINOGUANINE, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZO
 
 | | Crystal structure of folE riboswitch with Guanine | | Descriptor: | GUANINE, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZE
 
 | | Crystal structure of THF-II riboswitch with THF and soaked with Ir | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, IRIDIUM ION, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZM
 
 | | Crystal structure of folE riboswitch with DHN | | Descriptor: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZW
 
 | | Crystal structure of THF-II riboswitch with THF and soaked with Ir | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZQ
 
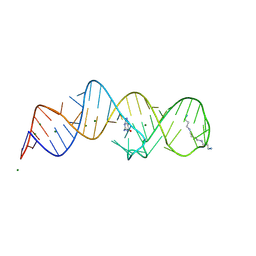 | | Crystal structure of folE riboswitch with 8-N Guanine | | Descriptor: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZP
 
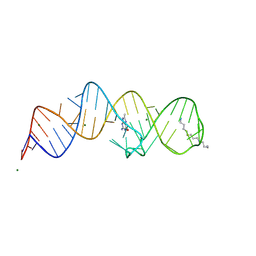 | | Crystal structure of folE riboswitch with 8-CH3 Guanine | | Descriptor: | 2-azanyl-8-methyl-1,9-dihydropurin-6-one, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZN
 
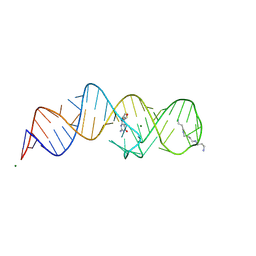 | | Crystal structure of folE riboswitch with BH4 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZL
 
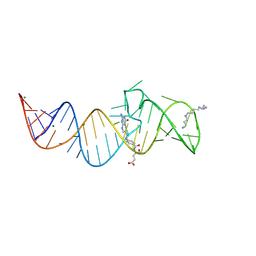 | | Crystal structure of folE riboswitch with DHF | | Descriptor: | DIHYDROFOLIC ACID, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
