3N4N
 
 | | Insights into the stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3' | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S. | | Deposit date: | 2010-05-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into the DNA stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido [2,3-d]pyrimidin-2-one.
Nucleic Acids Res., 2010
|
|
3LII
 
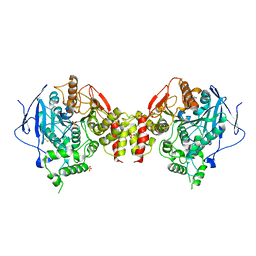 | | Recombinant human acetylcholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, SULFATE ION | | Authors: | Dvir, H, Rosenberry, T, Harel, M, Silman, I, Sussman, J. | | Deposit date: | 2010-01-25 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Acetylcholinesterase: From 3D structure to function.
Chem.Biol.Interact, 187, 2010
|
|
4Z2I
 
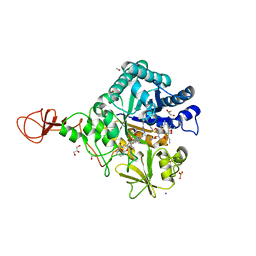 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 30 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
6W2U
 
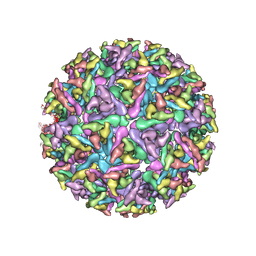 | |
6W09
 
 | |
6W1C
 
 | |
6VYV
 
 | |
4Z2G
 
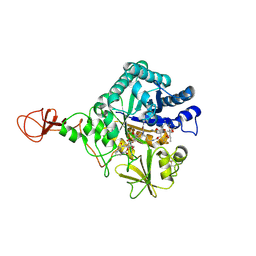 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 26 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {4-[N'-(methylcarbamoyl)carbamimidamido]butyl}carbamate, Chitinase B | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2L
 
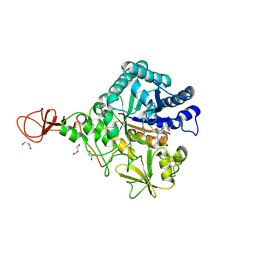 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 33 | | Descriptor: | (3R,4S,5S,6R,7R,9R,10S,11S,12R,13S,14R)-14-ethyl-7,10,12,13-tetrahydroxy-3,5,7,9,11,13-hexamethyl-2-oxo-6-(prop-2-yn-1-yloxy)oxacyclotetradecan-4-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2J
 
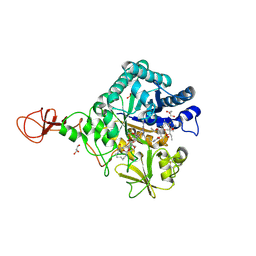 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 31 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {5-[N'-(methylcarbamoyl)carbamimidamido]pentyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2K
 
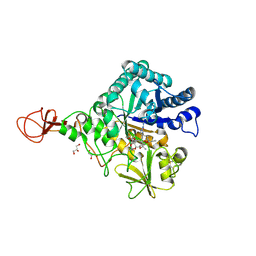 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 32 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {7-[N'-(methylcarbamoyl)carbamimidamido]heptyl}carbamate, Chitinase B, GLYCEROL | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2H
 
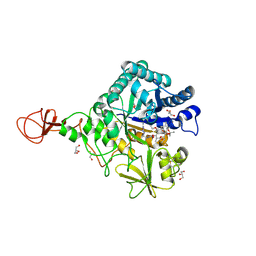 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 29 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {2-[N'-(methylcarbamoyl)carbamimidamido]ethyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
3L9B
 
 | | Crystal Structure of Rat Otoferlin C2A | | Descriptor: | MAGNESIUM ION, Otoferlin | | Authors: | Helfmann, S, Neumann, P. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the C2A domain of otoferlin reveals an unconventional top loop region.
J.Mol.Biol., 406, 2011
|
|
8FD2
 
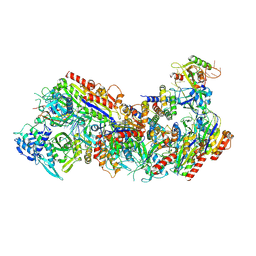 | | Cryo-EM structure of Cascade complex in type I-B CAST system | | Descriptor: | A Type I-B CRISPR-associated protein Cas5, RNA, Type I-B CRISPR-associated protein Cas11, ... | | Authors: | Chang, L, Wang, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector.
Cell, 186, 2023
|
|
8FCJ
 
 | |
8FCX
 
 | |
8FCW
 
 | |
8FF4
 
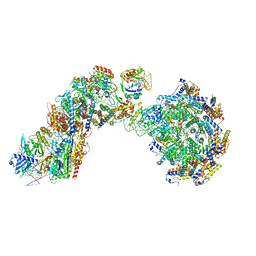 | |
8FCU
 
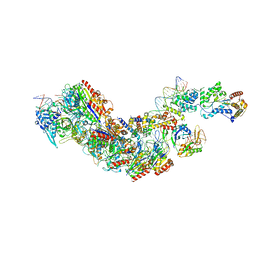 | |
8FD3
 
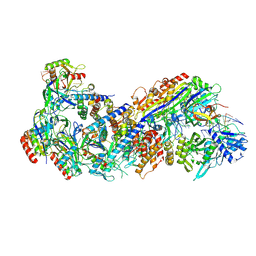 | |
8FCV
 
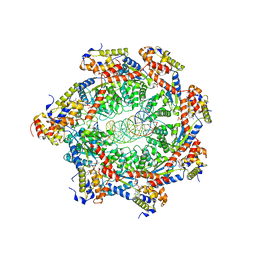 | |
8FF5
 
 | |
7T64
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T65
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the open state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2OBH
 
 | | Centrin-XPC peptide | | Descriptor: | CALCIUM ION, Centrin-2, DNA-repair protein complementing XP-C cells | | Authors: | Charbonnier, J.B. | | Deposit date: | 2006-12-19 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, thermodynamic, and cellular characterization of human centrin 2 interaction with xeroderma pigmentosum group C protein.
J.Mol.Biol., 373, 2007
|
|
