5TWM
 
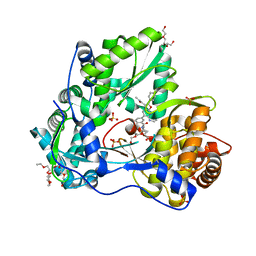 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS GENOTYPE 2A STRAIN JFH1 L30S NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA-dependent RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
6B30
 
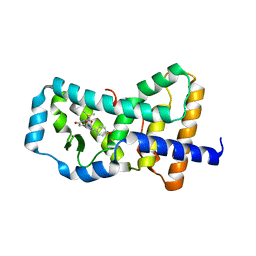 | | Structure of RORgt in complex with a novel inverse agonist 1 | | Descriptor: | N-[(1R)-1-(4-methoxyphenyl)-2-oxo-2-{[4-(trimethylsilyl)phenyl]amino}ethyl]-N-methyl-3-oxo-2,3-dihydro-1,2-oxazole-5-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of orally efficacious ROR gamma t inverse agonists, part 1: Identification of novel phenylglycinamides as lead scaffolds.
Bioorg. Med. Chem., 26, 2018
|
|
4TTY
 
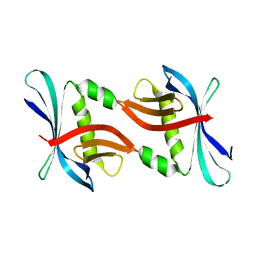 | |
5TWN
 
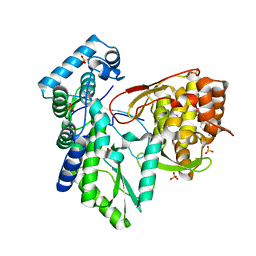 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA- DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA- DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
6E3E
 
 | | Structure of RORgt in complex with a novel inverse agonist. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(5R)-5-[(7-fluoro-1,1-dimethyl-2,3-dihydro-1H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5H)-yl]-5-oxopentanoic acid, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
6E3G
 
 | | Structure of RORgt in complex with a novel agonist. | | Descriptor: | (5R)-6-acetyl-2-methoxy-N-{4-[(2-methoxyphenyl)methoxy]phenyl}-5,6,7,8-tetrahydro-1,6-naphthyridine-5-carboxamide, 1,2-ETHANEDIOL, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
6B31
 
 | | Structure of RORgt in complex with a novel inverse agonist 2 | | Descriptor: | (3S)-N~1~-(3-chloro-4-cyanophenyl)-N~5~-(1,3-diethyl-2,4-dioxo-1,2,3,4-tetrahydroquinazolin-6-yl)-3-methylpentanediamide, Nuclear receptor ROR-gamma | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Identification of novel quinazolinedione derivatives as ROR gamma t inverse agonist.
Bioorg. Med. Chem., 26, 2018
|
|
1ABR
 
 | | CRYSTAL STRUCTURE OF ABRIN-A | | Descriptor: | ABRIN-A, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Tahirov, T.H, Lu, T.-H, Liaw, Y.-C, Chu, S.-C, Lin, J.-Y. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of abrin-a at 2.14 A.
J.Mol.Biol., 250, 1995
|
|
6B33
 
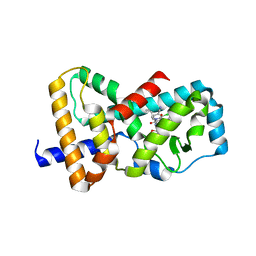 | | Structure of RORgt in complex with a novel inverse agonist 3 | | Descriptor: | (2R)-N~2~-(3-chloro-4-cyanophenyl)-N~4~-[3-(cyclopropylmethyl)-2,4-dioxo-1-(propan-2-yl)-1,2,3,4-tetrahydroquinazolin-6-yl]morpholine-2,4-dicarboxamide, Nuclear receptor ROR-gamma | | Authors: | Skene, R.J, Hoffman, I, Snell, G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design and Synthesis of Conformationally Constrained ROR gamma t Inverse Agonists.
Chemmedchem, 2019
|
|
7Q8I
 
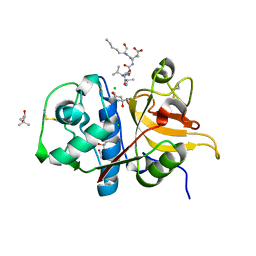 | | Peptide AVAEKQ in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AVAEKQ peptide, CHLORIDE ION, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q8O
 
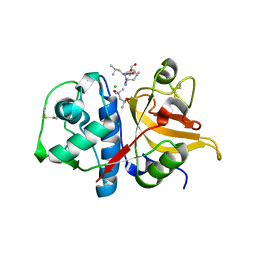 | | Peptide LLSGKE in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
1QM8
 
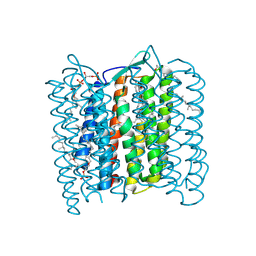 | | Structure of Bacteriorhodopsin at 100 K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Kamiya, N, Kouyama, T. | | Deposit date: | 1999-09-22 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Three-Dimensional Crystal of Bacteriorhodopsin Obtained by Successive Fusion of the Vesicular Assemblies.
J.Mol.Biol., 283, 1998
|
|
1SUJ
 
 | |
7PKI
 
 | |
2H6S
 
 | | Secreted aspartic proteinase (Sap) 3 from Candida albicans | | Descriptor: | Candidapepsin-3, ZINC ION | | Authors: | Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2006-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the secreted aspartic proteinase 3 from Candida albicans and its complex with pepstatin A.
Proteins, 68, 2007
|
|
3O0P
 
 | | Pilus-related Sortase C of Group B Streptococcus | | Descriptor: | Sortase family protein | | Authors: | Malito, E, Spraggon, G. | | Deposit date: | 2010-07-19 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus.
Faseb J., 25, 2011
|
|
6K33
 
 | | Structure of PSI-isiA supercomplex from Thermosynechococcus vulcanus | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Akita, F, Nagao, R, Kato, K, Shen, J.R, Miyazaki, N. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-20 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structure of a cyanobacterial photosystem I surrounded by octadecameric IsiA antenna proteins.
Commun Biol, 3, 2020
|
|
2H6T
 
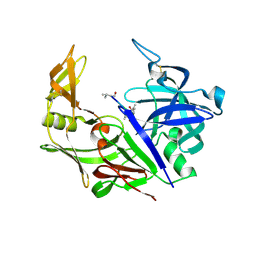 | | Secreted aspartic proteinase (Sap) 3 from Candida albicans complexed with pepstatin A | | Descriptor: | Candidapepsin-3, ZINC ION, pepstatin A | | Authors: | Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2006-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the secreted aspartic proteinase 3 from Candida albicans and its complex with pepstatin A.
Proteins, 68, 2007
|
|
5D5P
 
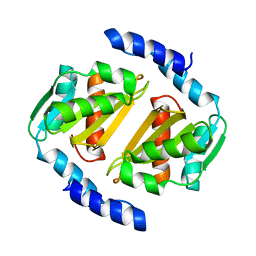 | | HcgB from Methanococcus maripaludis | | Descriptor: | HcgB | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards artificial methanogenesis: biosynthesis of the [Fe]-hydrogenase cofactor and characterization of the semi-synthetic hydrogenase.
Faraday Discuss., 198, 2017
|
|
3WIK
 
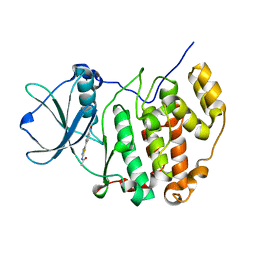 | | Crystal structure of the CK2alpha/compound10 complex | | Descriptor: | Casein kinase II subunit alpha, N-[5-(4-nitrophenyl)-1,3,4-thiadiazol-2-yl]acetamide | | Authors: | Kinoshita, T, Nakaniwa, T, Sekiguchi, Y, Nakanishi, I. | | Deposit date: | 2013-09-18 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Identification of protein kinase CK2 inhibitors using solvent dipole ordering virtual screening
To be Published
|
|
4K1C
 
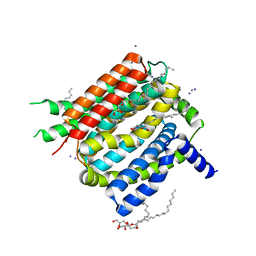 | | VCX1 Calcium/Proton Exchanger | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, IODIDE ION, ... | | Authors: | Waight, A.B, Pedersen, B.P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for alternating access of a eukaryotic calcium/proton exchanger.
Nature, 499, 2013
|
|
3WIL
 
 | | Crystal structure of the CK2alpha/compound3 complex | | Descriptor: | Casein kinase II subunit alpha, {[(2Z)-2-(3,4-dimethoxybenzylidene)-3-oxo-2,3-dihydro-1-benzofuran-6-yl]oxy}acetic acid | | Authors: | Kinoshita, T, Nakanishi, I. | | Deposit date: | 2013-09-18 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of protein kinase CK2 inhibitors using solvent dipole ordering virtual screening
To be Published
|
|
4KJT
 
 | | Structure of the L100F MUTANT OF DEHALOPEROXIDASE-HEMOGLOBIN A FROM AMPHITRITE ORNATA WITH OXYGEN | | Descriptor: | 1,2-ETHANEDIOL, Dehaloperoxidase A, OXYGEN MOLECULE, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-03 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Influence of heme environment structure on dioxygen affinity for the dual function Amphitrite ornata hemoglobin/dehaloperoxidase. Insights into the evolutional structure-function adaptations.
Arch.Biochem.Biophys., 545, 2014
|
|
2QZW
 
 | |
7VUH
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
