3NV4
 
 | |
3NV3
 
 | |
3NV2
 
 | |
3NV1
 
 | | Crystal structure of human galectin-9 C-terminal CRD | | Descriptor: | Galectin 9 short isoform variant, NICKEL (II) ION | | Authors: | Yoshida, H, Kamitori, S. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structures of human galectin-9 C-terminal domain in complexes with a biantennary oligosaccharide and sialyllactose
J.Biol.Chem., 285, 2010
|
|
8YEJ
 
 | | Cryo-EM structure of the channelrhodopsin GtCCR2 focused on the monomer | | Descriptor: | GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 84, 2024
|
|
8YEK
 
 | | Cryo-EM structure of the channelrhodopsin GtCCR2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 84, 2024
|
|
8YEL
 
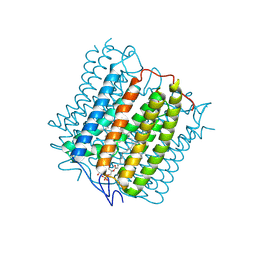 | | Cryo-EM structure of the channelrhodopsin GtCCR4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cation channel rhodopsin 4, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 84, 2024
|
|
1K37
 
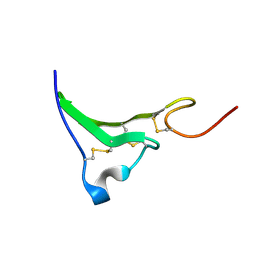 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
1K36
 
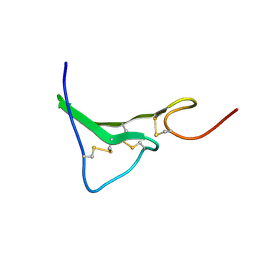 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
3WV6
 
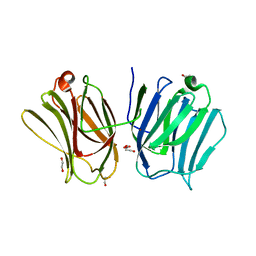 | |
5COK
 
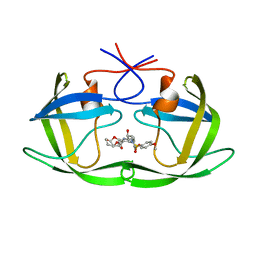 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-0476 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-097 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-(4-methoxyphenyl)butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5CON
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-015 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COO
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-085 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-1-(4-methoxyphenyl)-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
2E7A
 
 | |
4EN6
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4HL0
 
 | |
4EN8
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN9
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN7
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
1IRE
 
 | | Crystal Structure of Co-type nitrile hydratase from Pseudonocardia thermophila | | Descriptor: | COBALT (II) ION, Nitrile Hydratase | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Wakagi, T. | | Deposit date: | 2001-10-01 | | Release date: | 2002-10-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cobalt-containing nitrile hydratase.
Biochem.Biophys.Res.Commun., 288, 2001
|
|
3A4D
 
 | | Crystal structure of Human Transthyretin (wild-type) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of the Glutamic Acid 54 Residue in Transthyretin Stability and Thyroxine Binding
Biochemistry, 2009
|
|
3ACA
 
 | |
2ZJC
 
 | | TNFR1 selectve TNF mutant; R1-6 | | Descriptor: | GLYCEROL, Tumor necrosis factor | | Authors: | Mukai, Y, Yamagata, Y, Tsutsumi, Y. | | Deposit date: | 2008-03-05 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Function Relationship of Tumor Necrosis Factor (TNF) and Its Receptor Interaction Based on 3D Structural Analysis of a Fully Active TNFR1-Selective TNF Mutant
J.Mol.Biol., 385, 2009
|
|
3AC9
 
 | |
