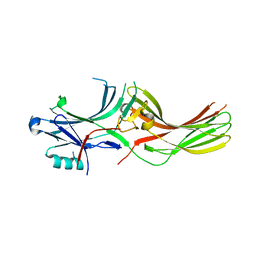
8AS4
 
 | |
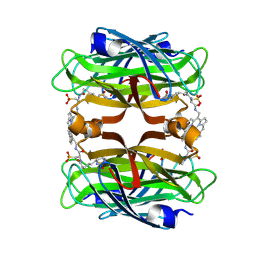
8TY0
 
 | |
8QEX
 
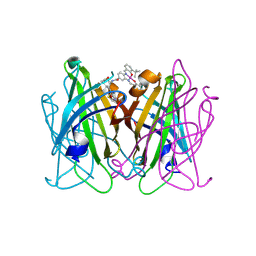 | |
8QQ3
 
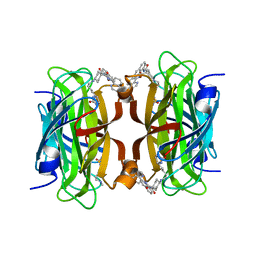 | | Streptavidin with a Ni-cofactor | | Descriptor: | 4-[4-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butylamino]-~{N}1,~{N}1'-di(quinolin-8-yl)cyclohexane-1,1-dicarboxamide, NICKEL (II) ION, Streptavidin | | Authors: | Zhang, K, Jakob, R.P, Ward, T.R. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An artificial nickel chlorinase based on the biotin-streptavidin technology.
Chem.Commun.(Camb.), 60, 2024
|
|
4QPT
 
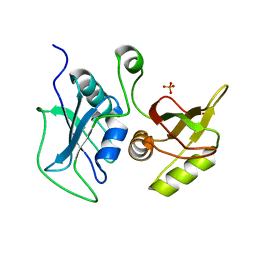 | | Structural Investigation of hnRNP L | | Descriptor: | Heterogenous nuclear ribonucleoprotein L, PHOSPHATE ION | | Authors: | Blatter, M, Allain, F.H.-T. | | Deposit date: | 2014-06-25 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | The Signature of the Five-Stranded vRRM Fold Defined by Functional, Structural and Computational Analysis of the hnRNP L Protein.
J.Mol.Biol., 427, 2015
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2MQL
 
 | | Structural Investigation of hnRNP L | | Descriptor: | Protein Hnrnpl | | Authors: | Blatter, M, Allain, F. | | Deposit date: | 2014-06-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Signature of the Five-Stranded vRRM Fold Defined by Functional, Structural and Computational Analysis of the hnRNP L Protein.
J.Mol.Biol., 427, 2015
|
|
2MQN
 
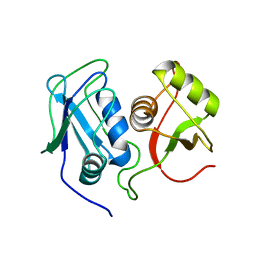 | | Structural Investigation of hnRNP L | | Descriptor: | Heterogenous nuclear ribonucleoprotein L | | Authors: | Blatter, M, Allain, F. | | Deposit date: | 2014-06-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Signature of the Five-Stranded vRRM Fold Defined by Functional, Structural and Computational Analysis of the hnRNP L Protein.
J.Mol.Biol., 427, 2015
|
|
2MQM
 
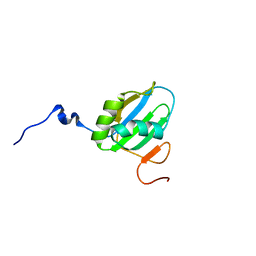 | | Structural Investigation of hnRNP L | | Descriptor: | Protein Hnrnpl | | Authors: | Blatter, M, Allain, F. | | Deposit date: | 2014-06-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Signature of the Five-Stranded vRRM Fold Defined by Functional, Structural and Computational Analysis of the hnRNP L Protein.
J.Mol.Biol., 427, 2015
|
|
9F15
 
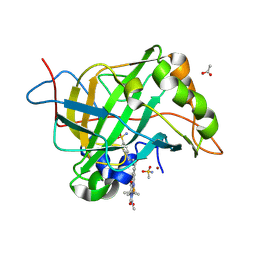 | | Carbonic anhydrase II variant with bound iron complex | | Descriptor: | 4,4,7,10,10-pentamethyl-3,6,8,11-tetrakis(oxidanylidene)-N-[2-(4-sulfamoylphenyl)ethyl]-2,5,7,9,12-pentazabicyclo[11.4.0]heptadeca-1(13),14,16-triene-15-carboxamide containing iron, ACETATE ION, Carbonic anhydrase 2, ... | | Authors: | Zhang, K, Jakob, R.P, Ward, T.R. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Directed Evolution of an Artificial Hydroxylase Based on a Thermostable Human Carbonic Anhydrase Protein
Acs Catalysis, 14, 2024
|
|
9F2E
 
 | | Carbonic anhydrase II variant with bound iron complex in space group C2 (ArPase) | | Descriptor: | 1,2-ETHANEDIOL, 4,4,7,10,10-pentamethyl-3,6,8,11-tetrakis(oxidanylidene)-N-[2-(4-sulfamoylphenyl)ethyl]-2,5,7,9,12-pentazabicyclo[11.4.0]heptadeca-1(13),14,16-triene-15-carboxamide containing iron, Carbonic anhydrase 2, ... | | Authors: | Jakob, R.P, Ward, T.R. | | Deposit date: | 2024-04-23 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Directed Evolution of an Artificial Hydroxylase Based on a Thermostable Human Carbonic Anhydrase Protein
Acs Catalysis, 14, 2024
|
|
9HGA
 
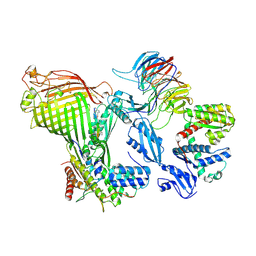 | | BAM-SurA-darobactin complex in the swing-out state | | Descriptor: | Chaperone SurA, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG8
 
 | | BAM-SurA complex in the swing-out state with BamC resolved | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG6
 
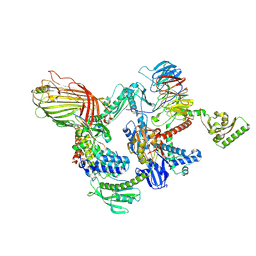 | | BAM-SurA complex in the swing-in state | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG9
 
 | | BAM-SurA-darobactin complex in the swing-in state | | Descriptor: | Chaperone SurA, Darobactin A, MAGNESIUM ION, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG7
 
 | | BAM-SurA complex in the swing-out state | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG5
 
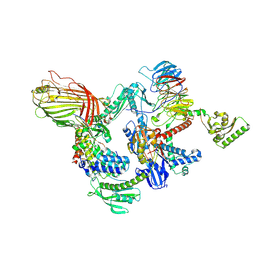 | | BAM-SurA complex in the swing-in state with full length BamC resolved | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9F2F
 
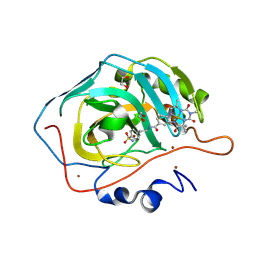 | | Carbonic anhydrase II variant with bound iron complex in space group R3 (ArPase) | | Descriptor: | 1,2-ETHANEDIOL, 4,4,7,10,10-pentamethyl-3,6,8,11-tetrakis(oxidanylidene)-N-[2-(4-sulfamoylphenyl)ethyl]-2,5,7,9,12-pentazabicyclo[11.4.0]heptadeca-1(13),14,16-triene-15-carboxamide containing iron, Carbonic anhydrase 2, ... | | Authors: | Jakob, R.P, Ward, T.R. | | Deposit date: | 2024-04-23 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Directed Evolution of an Artificial Hydroxylase Based on a Thermostable Human Carbonic Anhydrase Protein
Acs Catalysis, 14, 2024
|
|
3PZC
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with Coenzyme A | | Descriptor: | ACETATE ION, Amino acid--[acyl-carrier-protein] ligase 1, COENZYME A, ... | | Authors: | Weygand-Durasevic, I, Luic, M, Mocibob, M, Ivic, N, Subasic, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Recognition by Novel Family of Amino Acid:[Carrier Protein] Ligases
Croatica Chemica Acta, 84, 2011
|
|
4H2W
 
 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and AMP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4H2T
 
 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with cognate carrier protein and an analogue of glycyl adenylate | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 5'-O-(glycylsulfamoyl)adenosine, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4H2V
 
 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with glycylated carrier protein | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4H2Y
 
 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and ATP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4H2S
 
 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with cognate carrier protein and AMP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4H2X
 
 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and an analogue of glycyl adenylate | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 5'-O-(glycylsulfamoyl)adenosine, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
