8FLC
 
 | |
8FLD
 
 | |
5D51
 
 | | Krypton derivatization of an O2-tolerant membrane-bound [NiFe] hydrogenase reveals a hydrophobic gas tunnel network | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Frielingsdorf, S, van der Linden, P, von Stetten, D, Lenz, O, Carpentier, P, Scheerer, P. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Krypton Derivatization of an O2 -Tolerant Membrane-Bound [NiFe] Hydrogenase Reveals a Hydrophobic Tunnel Network for Gas Transport.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8FKX
 
 | |
8FKP
 
 | |
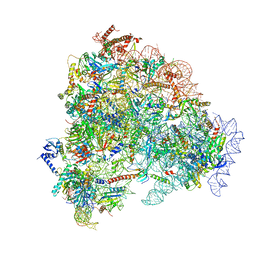
8FL0
 
 | |
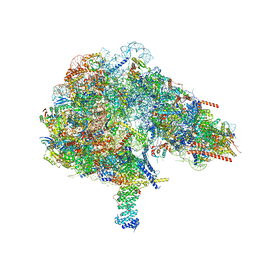
8FKW
 
 | |
5A6C
 
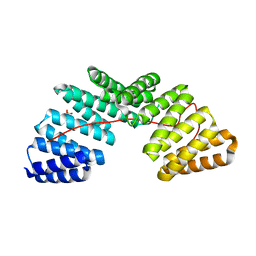 | | Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation | | Descriptor: | G-PROTEIN-SIGNALING MODULATOR 2, AFADIN, SULFATE ION | | Authors: | Carminati, M, Gallini, S, Pirovano, L, Alfieri, A, Bisi, S, Mapelli, M. | | Deposit date: | 2015-06-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Concomitant Binding of Afadin to Lgn and F-Actin Directs Planar Spindle Orientation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5WLC
 
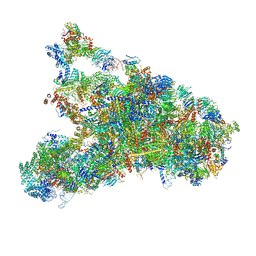 | | The complete structure of the small subunit processome | | Descriptor: | 18S pre-rRNA, 5' ETS, Bms1, ... | | Authors: | Barandun, J, Chaker-Margot, M, Hunziker, M, Klinge, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The complete structure of the small-subunit processome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7RHQ
 
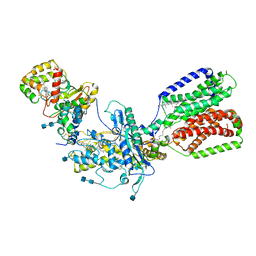 | | Cryo-EM structure of Xenopus Patched-1 in complex with GAS1 and Sonic Hedgehog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, P, Lian, T, Wierbowski, B, Garcia-Linares, S, Jiang, J, Salic, A. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
8CSQ
 
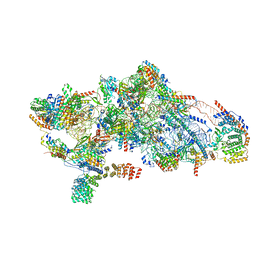 | | Human mitochondrial small subunit assembly intermediate (State B) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CSP
 
 | | Human mitochondrial small subunit assembly intermediate (State A) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
7Z57
 
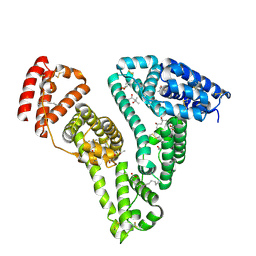 | | Crystal structure of Human Serum Albumin in complex with surfactant GenX (2,3,3,3-tetrafluoro-2-(heptafluoropropoxy) propanoate) | | Descriptor: | (2R)-2,3,3,3-tetrakis(fluoranyl)-2-[1,1,2,2,3,3,3-heptakis(fluoranyl)propoxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Albumin, ... | | Authors: | Liberi, S, Moro, G, Vascon, F, Linciano, S, De Toni, L, Angelin, A, Cendron, L. | | Deposit date: | 2022-03-08 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigation of the Interaction between Human Serum Albumin and Branched Short-Chain Perfluoroalkyl Compounds.
Chem.Res.Toxicol., 35, 2022
|
|
8CSR
 
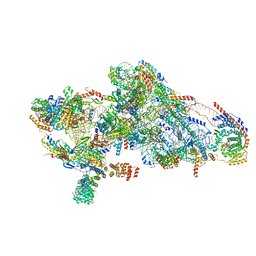 | | Human mitochondrial small subunit assembly intermediate (State C) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CSS
 
 | | Human mitochondrial small subunit assembly intermediate (State D) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8D8L
 
 | | Yeast mitochondrial small subunit assembly intermediate (State 3) | | Descriptor: | 15S ribosomal RNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Burnside, C, Harper, N, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CSU
 
 | | Human mitochondrial small subunit assembly intermediate (State C*) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CST
 
 | | Human mitochondrial small subunit assembly intermediate (State E) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8D8K
 
 | | Yeast mitochondrial small subunit assembly intermediate (State 2) | | Descriptor: | 15S ribosomal RNA, 37S ribosomal protein MRP1, mitochondrial, ... | | Authors: | Burnside, C, Harper, N.J, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8D8J
 
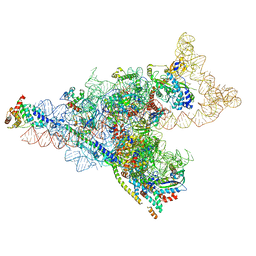 | | Yeast mitochondrial small subunit assembly intermediate (State 1) | | Descriptor: | 15S ribosomal RNA, 37S ribosomal protein MRP13, mitochondrial, ... | | Authors: | Burnside, C, Harper, N, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8E5T
 
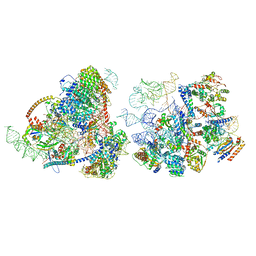 | | Yeast co-transcriptional Noc1-Noc2 RNP assembly checkpoint intermediate | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Sanghai, Z.A, Piwowarczyk, R, Vanden Broeck, A, Klinge, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A co-transcriptional ribosome assembly checkpoint controls nascent large ribosomal subunit maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EK9
 
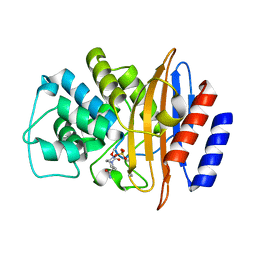 | | Crystal structure of the class A carbapenemase CRH-1 in complex with avibactam at 1.4 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Brunetti, F, Ghiglione, B, Guardabassi, L, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of CRH-1, a Carbapenemase from Chromobacterium haemolyticum Related to KPC beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
8EHU
 
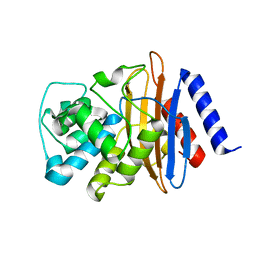 | | Crystal structure of the environmental CRH-1 class A carbapenemase at 1.1 Angstrom resolution | | Descriptor: | Beta-lactamase | | Authors: | Power, P, Brunetti, F, Ghiglione, B, Guardabassi, L, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Biochemical and Structural Characterization of CRH-1, a Carbapenemase from Chromobacterium haemolyticum Related to KPC beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
8EO5
 
 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8EHH
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Ghiglione, B, Bonomo, R.A, Rodriguez, M.M, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Biochemical and structural evidences of the activity of relebactam as inhibitor of the extended-spectrum beta-lactamase CTX-M-96.
To be published
|
|
