6OJ2
 
 | | Crystal structure of tRNA^ Ala(GGC) bound to the near-cognate 70S A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-04-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OPE
 
 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to near-cognate 70S A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-04-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ORD
 
 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to cognate 70S A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8WXL
 
 | | Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
6OF6
 
 | | Crystal structure of tRNA^ Ala(GGC) bound to cognate 70S A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6K60
 
 | | Structural and functional basis for HLA-G isoform recognition of immune checkpoint receptor LILRBs | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain G, ... | | Authors: | Kuroki, K, Matsubara, H, Kanda, R, Miyashita, N, Shiroishi, M, Fukunaga, Y, Kamishikiryo, J, Fukunaga, A, Hirose, K, Sugita, Y, Kita, S, Ose, T, Maenaka, K. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms.
J Immunol., 203, 2019
|
|
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
3ASE
 
 | | Crystal Structure of Zinc myoglobin soaked with Ru3O cluster | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING ZN, SULFATE ION, ... | | Authors: | Koshiyama, T, Shirai, M, Hikage, T, Tabe, H, Tanaka, K, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Post-Crystal Engineering of Zinc-Substituted Myoglobin to Construct a Long-Lived Photoinduced Charge-Separation System
Angew.Chem.Int.Ed.Engl., 2011
|
|
6KZA
 
 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | Descriptor: | DNA replication protein DnaC, Replicative DNA helicase | | Authors: | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D48
 
 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | Descriptor: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | Descriptor: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
1DF8
 
 | | S45A MUTANT OF STREPTAVIDIN IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Hyre, D.E, Le Trong, I, Freitag, S, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 1999-11-18 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Ser45 plays an important role in managing both the equilibrium and transition state energetics of the streptavidin-biotin system.
Protein Sci., 9, 2000
|
|
1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
2E4T
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2006-12-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EC5
 
 | |
2EEX
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-02-19 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EBF
 
 | |
2EJ1
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2E0P
 
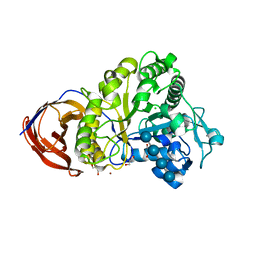 | | The crystal structure of Cel44A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2006-10-11 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EBH
 
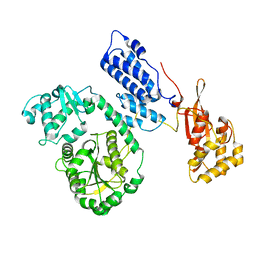 | |
2EO7
 
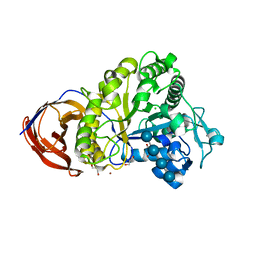 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitao, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-29 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EQD
 
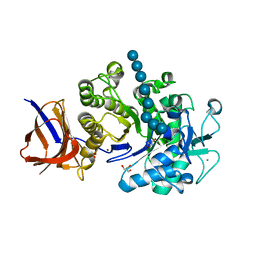 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-30 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2PSM
 
 | | Crystal structure of Interleukin 15 in complex with Interleukin 15 receptor alpha | | Descriptor: | BENZAMIDINE, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Olsen, S.K, Murayama, K, Kishishita, S, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Ota, N, Kanagawa, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Interleukin-15{middle dot}Interleukin-15 Receptor {alpha} Complex: INSIGHTS INTO TRANS AND CIS PRESENTATION
J.Biol.Chem., 282, 2007
|
|
