2WW5
 
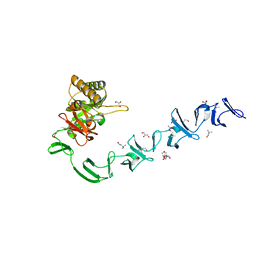 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L, Menendez, M. | | Deposit date: | 2009-10-21 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2X0K
 
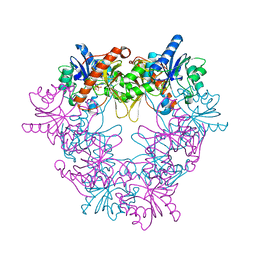 | | Crystal structure of modular FAD synthetase from Corynebacterium ammoniagenes | | Descriptor: | PYROPHOSPHATE, RIBOFLAVIN BIOSYNTHESIS PROTEIN RIBF, SULFATE ION | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Medina, M, Frago, S. | | Deposit date: | 2009-12-15 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligomeric State in the Crystal Structure of Modular Fad Synthetase Provides Insights Into its Sequential Catalysis in Prokaryotes
J.Mol.Biol., 400, 2010
|
|
2WWD
 
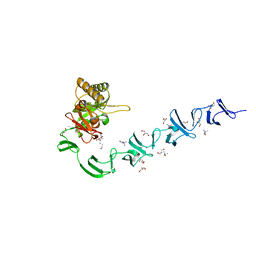 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALANINE, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2YP6
 
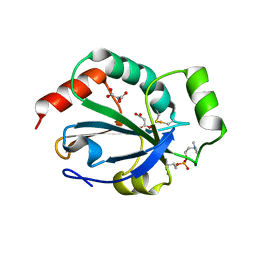 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_1000 (Etrx2) from Streptococcus pneumoniae strain TIGR4 in complex with Cyclofos 3 TM | | Descriptor: | 3-Cyclohexyl-1-Propylphosphocholine, MALONATE ION, THIOREDOXIN FAMILY PROTEIN | | Authors: | Bartual, S.G, Shalek, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Molecular Architecture of Streptococcus Pneumoniae Surface Thioredoxin-Fold Lipoproteins Crucial for Extracellular Oxidative Stress Resistance and Maintenance of Virulence.
Embo Mol.Med., 5, 2013
|
|
1O71
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with glycerol | | Descriptor: | GLYCEROL, STICHOLYSIN II | | Authors: | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-10-23 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
1O72
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with phosphorylcholine | | Descriptor: | PHOSPHOCHOLINE, STICHOLYSIN II | | Authors: | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-10-23 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
1GZ7
 
 | | Crystal structure of the closed state of lipase 2 from Candida rugosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LIPASE 2 | | Authors: | Mancheno, J.M, Hermoso, J.A. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights Into the Lipase/Esterase Behavior in the Candida Rugosa Lipases Family: Crystal Structure of the Lipase 2 Isoenzyme at 1.97A Resolution
J.Mol.Biol., 332, 2003
|
|
1GWY
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II | | Descriptor: | STICHOLYSIN II, SULFATE ION | | Authors: | Mancheno, J.M, Martin-Benito, J, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-03-26 | | Release date: | 2003-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
4BJ4
 
 | | Structure of Pseudomonas aeruginosa amidase Ampdh2 | | Descriptor: | AMPDH2, CITRATE ANION | | Authors: | Martinez-Caballero, C.S, Carrasco-Lopez, C, Artola-Recolons, C, Hermoso, J.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4BF5
 
 | |
4BL2
 
 | |
4BHY
 
 | | Structure of alanine racemase from Aeromonas hydrophila | | Descriptor: | ALANINE RACEMASE | | Authors: | Otero, L.H, Carrasco-Lopez, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-04-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for the Broad Specificity of a New Family of Amino-Acid Racemases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CFP
 
 | | Crystal structure of MltC in complex with tetrasaccharide at 2.15 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Bernardo-Garcia, N, Artola-Recolons, C, Hermoso, J.A. | | Deposit date: | 2013-11-19 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4CFO
 
 | | Structure of Lytic Transglycosylase MltC from Escherichia coli in complex with tetrasaccharide at 2.9 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranoside, MLTC | | Authors: | Artola-Recolons, C, Bernardo-Garcia, N, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-11-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4BXD
 
 | |
4BXJ
 
 | |
4CAJ
 
 | | Crystallographic structure of the mouse SIGN-R1 CRD domain in complex with sialic acid | | Descriptor: | CALCIUM ION, CD209 ANTIGEN-LIKE PROTEIN B, CHLORIDE ION, ... | | Authors: | Silva-Martin, N, Bartual, S.G, Hermoso, J.A. | | Deposit date: | 2013-10-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
4BPA
 
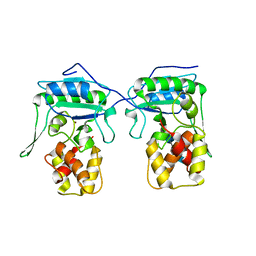 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with NAG-NAM-NAG-NAM tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, AMPDH2, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E, Lastochkin, E, Zhang, W, Hellman, L, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4BXE
 
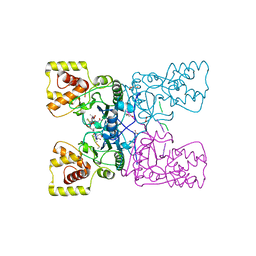 | |
4BUR
 
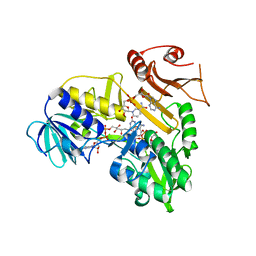 | | Crystal structure of the reduced human Apoptosis inducing factor complexed with NAD | | Descriptor: | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
4BV6
 
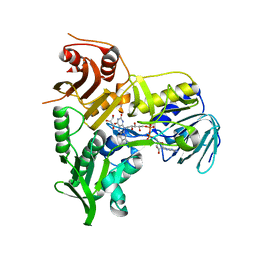 | | Refined crystal structure of the human Apoptosis inducing factor | | Descriptor: | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
4C5F
 
 | |
4BE4
 
 | | Closed conformation of O. piceae sterol esterase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gutierrez-Fernandez, J, Vaquero, M.E, Prieto, A, Barriuso, J, Gonzalez, M.J, Hermoso, J.A. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release.
J.Struct.Biol., 187, 2014
|
|
4BEU
 
 | |
4BL3
 
 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
