5IPQ
 
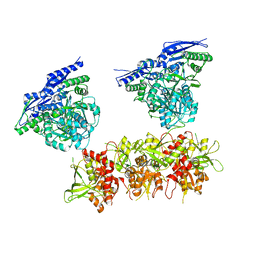 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 2 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IOU
 
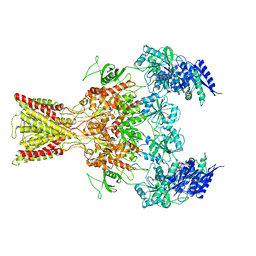 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine-bound conformation | | Descriptor: | GLUTAMIC ACID, GLYCINE, Ionotropic glutamate receptor subunit NR2B, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPU
 
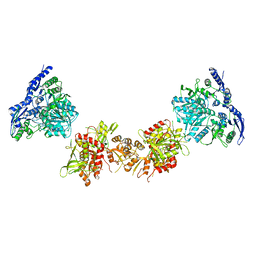 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 6 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5I6X
 
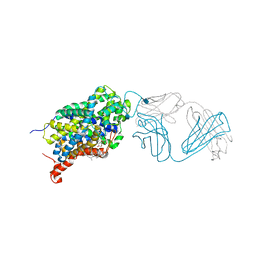 | | X-ray structure of the ts3 human serotonin transporter complexed with paroxetine at the central site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, heavy chain, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
3IJ4
 
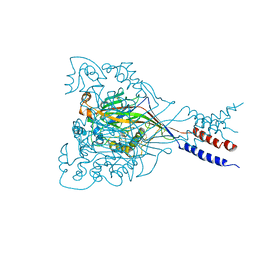 | |
5I66
 
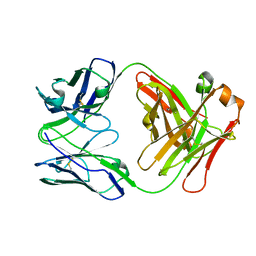 | | X-ray structure of the Fab fragment of 8B6, a murine monoclonal antibody specific for the human serotonin transporter | | Descriptor: | 8B6 antibody, heavy chain, light chain | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
5I75
 
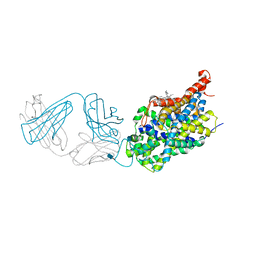 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central site and Br-citalopram at the allosteric site | | Descriptor: | (1S)-1-(4-bromophenyl)-1-[3-(dimethylamino)propyl]-1,3-dihydro-2-benzofuran-5-carbonitrile, (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
5IPS
 
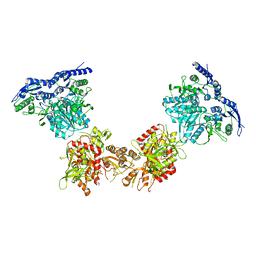 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 4 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
3H5W
 
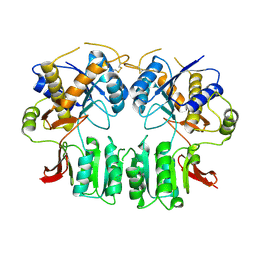 | | Crystal structure of the GluR2-ATD in space group P212121 without solvent | | Descriptor: | Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
3H5V
 
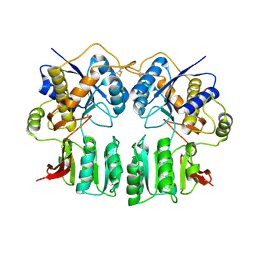 | | Crystal structure of the GluR2-ATD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
1MY1
 
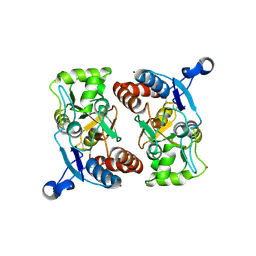 | | crystal titration experiments (AMPA co-crystals soaked in 10 nM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
1MQG
 
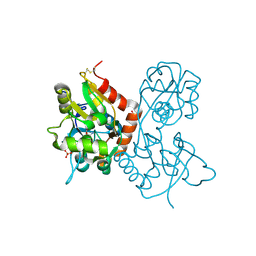 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MXX
 
 | | crystal titration experiments (AMPA co-crystals soaked in 100 uM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
1MY3
 
 | | crystal structure of glutamate receptor ligand-binding core in complex with bromo-willardiine in the Zn crystal form | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core:
Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
1MQJ
 
 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution | | Descriptor: | 2-AMINO-3-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ZINC ION, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
3JAF
 
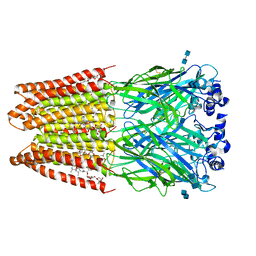 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine/ivermectin-bound state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
3TT3
 
 | |
3TU0
 
 | |
3TT1
 
 | |
3USI
 
 | |
3USK
 
 | |
3USL
 
 | | Crystal Structure of LeuT bound to L-selenomethionine in space group C2 from lipid bicelles | | Descriptor: | ACETATE ION, IODIDE ION, PHOSPHOCHOLINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3USO
 
 | |
3USJ
 
 | |
3USM
 
 | | Crystal Structure of LeuT bound to L-selenomethionine in space group C2 from lipid bicelles (collected at 1.2 A) | | Descriptor: | IODIDE ION, PHOSPHOCHOLINE, SELENOMETHIONINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
