6QJF
 
 | |
8SHR
 
 | | Crystal Structure of PRMT3 with Compound YD1-214 | | Descriptor: | 5'-S-[2-(phenylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-214
To be published
|
|
6QJL
 
 | |
8SIO
 
 | | Crystal structure of PRMT3 with YD1-66 | | Descriptor: | 5'-S-{3-[N'-(4'-chloro[1,1'-biphenyl]-3-yl)carbamimidamido]propyl}-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PRMT3 with YD1-66
To be published
|
|
8R9R
 
 | |
6QJC
 
 | |
6QL0
 
 | |
6QKB
 
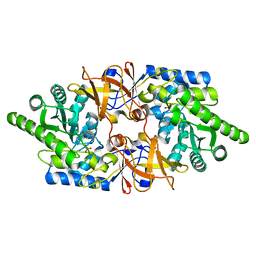 | | Crystal structure of the beta-hydroxyaspartate aldolase of Paracoccus denitrificans | | Descriptor: | D-3-hydroxyaspartate aldolase, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zarzycki, J, Schada von Borzyskowski, L, Gilardet, A, Erb, T.J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Marine Proteobacteria metabolize glycolate via the beta-hydroxyaspartate cycle.
Nature, 575, 2019
|
|
6QL1
 
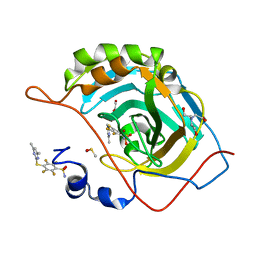 | | Crystal structure of chimeric carbonic anhydrase VI with 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-01-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Engineered Carbonic Anhydrase VI-Mimic Enzyme Switched the Structure and Affinities of Inhibitors.
Sci Rep, 9, 2019
|
|
8SH7
 
 | |
6Q2Z
 
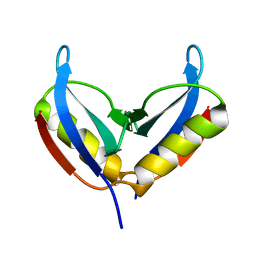 | | NMR solution structure of the HVO_2922 protein from Haloferax volcanii | | Descriptor: | UPF0339 family protein | | Authors: | Kubatova, N, Jonker, H.R.A, Saxena, K, Richter, C, Marchfelder, A, Schwalbe, H. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Small Protein HVO_2922 from Haloferax volcanii.
Chembiochem, 21, 2020
|
|
8R5K
 
 | |
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6QQ8
 
 | |
6QQD
 
 | |
6QQA
 
 | |
6QQF
 
 | |
6QQP
 
 | | Aplysia californica AChBP in complex with 2-Fluoro-(carbamoylpyridinyl)deschloroepibatidine analogue (2) | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(1~{R},2~{R},4~{S})-7-azabicyclo[2.2.1]heptan-2-yl]-2-fluoranyl-pyridin-3-yl]pyridine-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bueno, R.V, Davis, S, Dawson, A, Hunter, W.N. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interactions between 2'-fluoro-(carbamoylpyridinyl)deschloroepibatidine analogues and acetylcholine-binding protein inform on potent antagonist activity against nicotinic receptors
Acta Crystallogr.,Sect.D, 78, 2022
|
|
6QSN
 
 | | Crystal structure of Yellow fever virus polymerase NS5A | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Boura, E, Dubankova, A. | | Deposit date: | 2019-02-21 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses.
Antiviral Res., 169, 2019
|
|
4TXN
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, Uridine phosphorylase | | Authors: | Marinho, A, Torini, J, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
6QQB
 
 | |
4TXH
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in APO form | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Torini, J, Romanello, L, Marinho, A, Brandao-Neto, J, Cassago, A, DeMarco, R, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
6QQH
 
 | |
8SFG
 
 | | Crystal Structure of the Open Unbound Catalytically Inactive Makes Caterpillars Floppy-like (MCF) Effector from Vibrio vulnificus CMCP6 | | Descriptor: | Autotransporter adhesin, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Herrera, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-04-11 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Open Unbound Catalytically Inactive Makes Caterpillars Floppy-like (MCF) Effector from Vibrio vulnificus CMCP6.
To Be Published
|
|
4TXJ
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with thymidine | | Descriptor: | SULFATE ION, THYMIDINE, Uridine phosphorylase | | Authors: | Torini, J, Marinho, A, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
