6F5V
 
 | | Crystal structure of the prephenate aminotransferase from Arabidopsis thaliana | | Descriptor: | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, CITRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Robin, A, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-03 | | Release date: | 2019-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
6F1X
 
 | | Complex between MTH1 and compound 7 (a 7-azaindole-2-amide derivative) | | Descriptor: | 4-(3-chlorophenyl)-~{N}-ethyl-1~{H}-pyrrolo[2,3-b]pyridine-2-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6F2M
 
 | | Structure of the bacteriophage T5 distal tail protein pb9 co-crystallized with 10mM Tb-Xo4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Distal tail protein, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Breyton, C, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6ETB
 
 | | Aerobic S262Y mutation of E. coli FLRD core | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Romao, C.V, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | A tyrosine mutation in E. coli flavodiiron protein increases radiation sensitivity in the crystal structure
To Be Published
|
|
6F67
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with 3,4-Dihydroxybenzophenone | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, [3,4-bis(oxidanyl)phenyl]-phenyl-methanone, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
6EVS
 
 | | Characterization of 2-deoxyribosyltransferase from psychrotolerant bacterium Bacillus psychrosaccharolyticus: a suitable biocatalyst for the industrial synthesis of antiviral and antitumoral nucleosides | | Descriptor: | N-deoxyribosyltransferase | | Authors: | Fresco-Tabohada, A, Fernandez-Lucas, J, Acebal, C, Arroyo, M, Ramon, F, Mancheno, J.M, de la Mata, I. | | Deposit date: | 2017-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2'-Deoxyribosyltransferase from Bacillus psychrosaccharolyticus: A Mesophilic-Like Biocatalyst for the Synthesis of Modified Nucleosides from a Psychrotolerant Bacterium
Catalysts, 2019
|
|
6F93
 
 | | Helicobacter pylori serine hydroxymethyl transferase in apo form | | Descriptor: | Serine hydroxymethyltransferase | | Authors: | Sodolescu, A, Dian, C, Terradot, L, Bouzhir-Sima, L, Lestini, R, Myllykallio, H, Skouloubris, S, Liebl, U. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insight into serine hydroxymethyltransferase from Helicobacter pylori.
PLoS ONE, 13, 2018
|
|
6EZM
 
 | | Imidazoleglycerol-phosphate dehydratase from Saccharomyces cerevisiae | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2R)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6F0X
 
 | | Cryo-EM structure of TRIP13 in complex with ATP gamma S, p31comet, C-Mad2 and Cdc20 | | Descriptor: | Cell division cycle protein 20 homolog, MAD2L1-binding protein, Mitotic spindle assembly checkpoint protein MAD2A, ... | | Authors: | Alfieri, C, Chang, L, Barford, D. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13.
Nature, 559, 2018
|
|
6F23
 
 | | Complex between MTH1 and compound 16 (a 4-amino-7-azaindole derivative) | | Descriptor: | 4-[(2~{R})-2-phenylpyrrolidin-1-yl]-1~{H}-pyrrolo[2,3-b]pyridine, 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, ... | | Authors: | Viklund, J, Tresaugues, L, Talagas, A, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6FD2
 
 | | Radical SAM 1,2-diol dehydratase AprD4 in complex with its substrate paromamine | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Liu, W.Q, Amara, P, Mouesca, J.M, Ji, X, Renoux, O, Martin, L, Zhang, C, Zhang, Q, Nicolet, Y. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 1,2-Diol Dehydration by the Radical SAM Enzyme AprD4: A Matter of Proton Circulation and Substrate Flexibility.
J. Am. Chem. Soc., 140, 2018
|
|
6F35
 
 | | Crystal structure of the aspartate aminotranferase from Rhizobium meliloti | | Descriptor: | ACETATE ION, Aspartate aminotransferase B, GLYCEROL, ... | | Authors: | Cobessi, D, Graindorge, M, Giustini, C, Matringe, M. | | Deposit date: | 2017-11-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
6F43
 
 | | Crystal structure of apo form of Glutathione transferase Omega 3S from Trametes versicolor | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
9FP5
 
 | | Coxsackievirus A9 bound with CL213. | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
6F55
 
 | | Complex structure of PACSIN SH3 domain and TRPV4 proline rich region | | Descriptor: | PACSIN 3, PRR | | Authors: | Glogowski, N.A, Goretzki, B, Diehl, E, Duchardt-Ferner, E, Hacker, C, Hellmich, U.A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2.
Structure, 26, 2018
|
|
7A5X
 
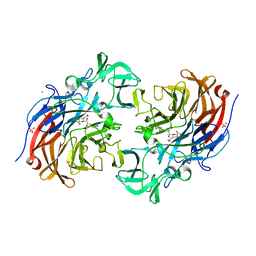 | | Two copies of the catalytic domain of NanA sialidase from Streptococcus pneumoniae juxtaposed in the P212121 space group, in complex with DANA derivatized with a PEG linker on the glycerol group. | | Descriptor: | (4~{S},5~{R},6~{R})-5-acetamido-6-[(1~{R},2~{S})-3-[[1-(2-ethoxyethyl)-1,2,3-triazol-4-yl]methylsulfanyl]-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bridot, C, Bouckaert, J. | | Deposit date: | 2020-08-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polyvalent Transition-State Analogues of Sialyl Substrates Strongly Inhibit Bacterial Sialidases*.
Chemistry, 27, 2021
|
|
6EYP
 
 | | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Prokofev, I.I, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A
To Be Published
|
|
6EXO
 
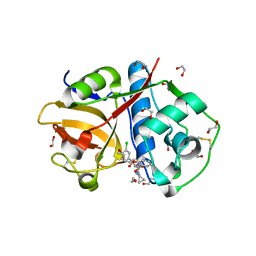 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6F1Y
 
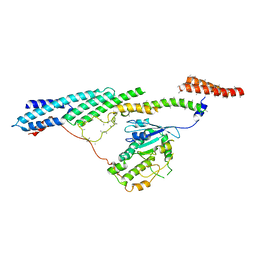 | | Dynein light intermediate chain region of the dynein tail/dynactin/BICDR1 complex | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1,Dynein heavy chain, Cytoplasmic dynein 1 light intermediate chain 2 | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
6F2T
 
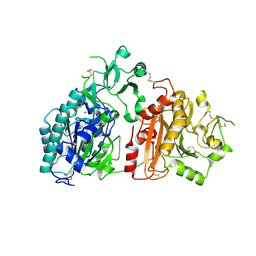 | | Crystal structure of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dohler, C, Zebisch, M, Strater, N. | | Deposit date: | 2017-11-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and substrate binding mode of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3).
Sci Rep, 8, 2018
|
|
6F64
 
 | | Crystal structure of the SYCP1 C-terminal back-to-back assembly | | Descriptor: | ACETATE ION, Synaptonemal complex protein 1 | | Authors: | Dunce, J.M, Millan, C, Uson, I, Davies, O.R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural basis of meiotic chromosome synapsis through SYCP1 self-assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6F4K
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with hexyl-glutathione | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
6F51
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with glutathionyl-phenylacetophenone | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
6FAD
 
 | | SR protein kinase 1 (SRPK1) in complex with the RGG-box of HSV1 ICP27 | | Descriptor: | PHOSPHATE ION, SRSF protein kinase 1, mRNA export factor | | Authors: | Tunnicliffe, R.B, Levy, C, Mould, A.P, Mckenzie, E.A, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2017-12-15 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Molecular Mechanism of SR Protein Kinase 1 Inhibition by the Herpes Virus Protein ICP27.
Mbio, 10, 2019
|
|
6F77
 
 | | Crystal structure of the prephenate aminotransferase from Rhizobium meliloti | | Descriptor: | Aspartate aminotransferase A, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cobessi, D, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
