8EM4
 
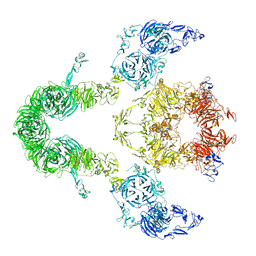 | | Cryo-EM structure of LRP2 at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Brasch, J, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
1GD6
 
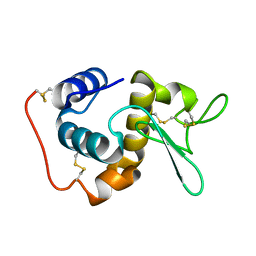 | | STRUCTURE OF THE BOMBYX MORI LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Matsuura, A, Aizawa, T, Yao, M, Kawano, K, Tanaka, I, Nitta, K. | | Deposit date: | 2000-09-19 | | Release date: | 2001-03-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of an insect lysozyme exhibiting catalytic efficiency at low temperatures.
Biochemistry, 41, 2002
|
|
8F4T
 
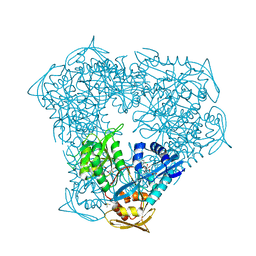 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with proguanil | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Punetha, A, Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
1LBJ
 
 | |
8F4Z
 
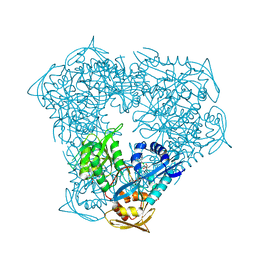 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with chloroquine | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-acetyltransferase Eis, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F58
 
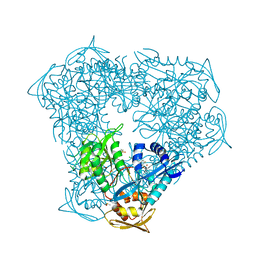 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with inhibitor SGT1616 | | Descriptor: | (1E)-1-[(4-chlorophenyl)methylidene]-2-(4-fluorophenyl)hydrazine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
1GGV
 
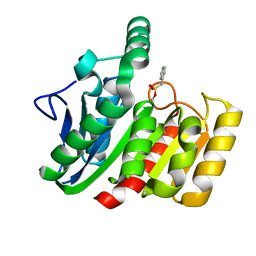 | | CRYSTAL STRUCTURE OF THE C123S MUTANT OF DIENELACTONE HYDROLASE (DLH) BOUND WITH THE PMS MOIETY OF THE PROTEASE INHIBITOR, PHENYLMETHYLSULFONYL FLUORIDE (PMSF) | | Descriptor: | DIENELACTONE HYDROLASE | | Authors: | Robinson, A, Edwards, K.J, Carr, P.D, Barton, J.D, Ewart, G.D, Ollis, D.L. | | Deposit date: | 2000-09-27 | | Release date: | 2000-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the C123S mutant of dienelactone hydrolase (DLH) bound with the PMS moiety of the protease inhibitor phenylmethylsulfonyl fluoride (PMSF).
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8F4U
 
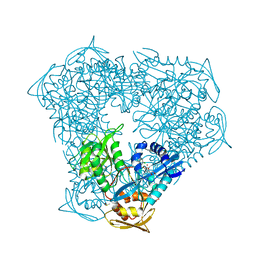 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with azelastine | | Descriptor: | 4-[(4-chlorophenyl)methyl]-2-[(4S)-1-methylazepan-4-yl]phthalazin-1(2H)-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
1GJ6
 
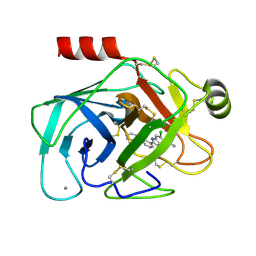 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GNG
 
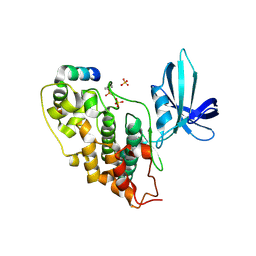 | | Glycogen synthase kinase-3 beta (GSK3) complex with FRATtide peptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FRATTIDE, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Bax, B, Carter, P.S, Lewis, C, Guy, A.R, Bridges, A, Tanner, R, Pettman, G, Mannix, C, Culbert, A.A, Brown, M.J.B, Smith, D.G, Reith, A.D. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of Phosphorylated Gsk-3Beta Complexed with a Peptide, Frattide, that Inhibits Beta-Catenin Phosphorylation
Structure, 9, 2001
|
|
8F51
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with mefloquine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
1GJA
 
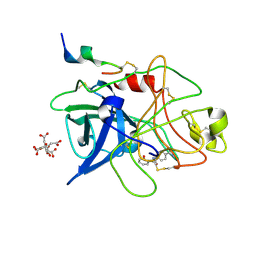 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | CITRIC ACID, N-(4-CARBAMIMIDOYL-PHENYL)-2-HYDROXY-BENZAMIDE, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJ8
 
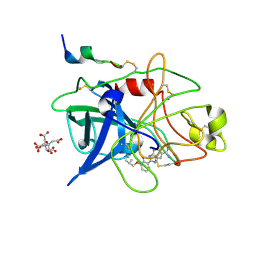 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 6-FLUORO-2-(2-HYDROXY-3-ISOBUTOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
2O0W
 
 | |
1GN0
 
 | | Escherichia coli GlpE sulfurtransferase soaked with KCN | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-10-01 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
1GJ5
 
 | | SELECTIVITY AT S1, H2O DISPLACEMENT, UPA, TPA, SER190/ALA190 PROTEASE, STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJU
 
 | | Maltosyltransferase from Thermotoga maritima | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-02 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
1GJX
 
 | | Solution structure of the lipoyl domain of the chimeric dihydrolipoyl dehydrogenase P64K from Neisseria meningitidis | | Descriptor: | PYRUVATE DEHYDROGENASE | | Authors: | Tozawa, K, Broadhurst, R.W, Raine, A.R.C, Fuller, C, Alvarez, A, Guillen, G, Padron, G, Perham, R.N. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Lipoyl Domain of the Chimeric Dihydrolipoyl Dehydrogenase P64K from Neisseria Meningitidis
Eur.J.Biochem., 268, 2001
|
|
1XZG
 
 | |
1L9G
 
 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
1GMX
 
 | | Escherichia coli GlpE sulfurtransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
1I5K
 
 | | STRUCTURE AND BINDING DETERMINANTS OF THE RECOMBINANT KRINGLE-2 DOMAIN OF HUMAN PLASMINOGEN TO AN INTERNAL PEPTIDE FROM A GROUP A STREPTOCOCCAL SURFACE PROTEIN | | Descriptor: | M PROTEIN, PLASMINOGEN | | Authors: | Rios-Steiner, J.L, Schenone, M, Mochalkin, I, Tulinsky, A, Castellino, F.J. | | Deposit date: | 2001-02-27 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein.
J.Mol.Biol., 308, 2001
|
|
1PXW
 
 | | Crystal structure of L7Ae sRNP core protein from Pyrococcus abyssii | | Descriptor: | LSU ribosomal protein L7AE | | Authors: | Charron, C, Manival, X, Charpentier, B, Branlant, C, Aubry, A. | | Deposit date: | 2003-07-07 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Purification, crystallization and preliminary X-ray diffraction data of L7Ae sRNP core protein from Pyrococcus abyssii.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8EZ1
 
 | | Human Ornithine Aminotransferase (hOAT) co-crystallized with its inactivator 3-Amino-4-fluorocyclopentenecarboxylic Acid | | Descriptor: | (1R,3S,4Z)-3-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-4-iminocyclopentane-1-carboxylic acid, (3E,4E)-4-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-3-iminocyclopent-1-ene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Shen, S, Silverman, R, Liu, D. | | Deposit date: | 2022-10-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Mechanistic Basis for the Inactivation of Human Ornithine Aminotransferase by (3 S ,4 S )-3-Amino-4-fluorocyclopentenecarboxylic Acid.
Molecules, 28, 2023
|
|
2NVO
 
 | |
