8EEQ
 
 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-09-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
4O27
 
 | | Crystal structure of MST3-MO25 complex with WIF motif | | Descriptor: | 5-mer peptide from serine/threonine-protein kinase 24, ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, ... | | Authors: | Hao, Q, Feng, M, Zhou, Z.C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.185 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
4ZMJ
 
 | | Crystal Structure of Ligand-Free BG505 SOSIP.664 HIV-1 Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2015-05-04 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2LJ1
 
 | | The third SH3 domain of R85FL with ataxin-7 PRR | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
5C77
 
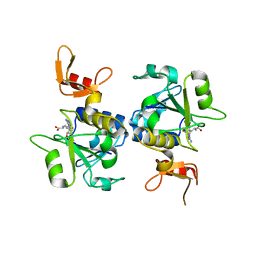 | | A novel protein arginine methyltransferase | | Descriptor: | Protein arginine N-methyltransferase SFM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lv, F, Zhang, T, Ding, J. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Sfm1 functioning as a protein arginine methyltransferase.
Cell Discov, 1, 2015
|
|
5C74
 
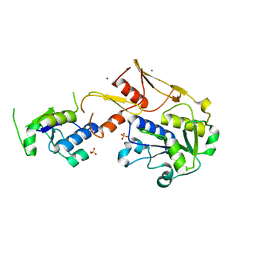 | | Structure of a novel protein arginine methyltransferase | | Descriptor: | NICKEL (II) ION, Protein arginine N-methyltransferase SFM1, SULFATE ION | | Authors: | Lv, F, Ding, J. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for Sfm1 functioning as a protein arginine methyltransferase.
Cell Discov, 1, 2015
|
|
2LJ0
 
 | | The third SH3 domain of R85FL | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
6IUR
 
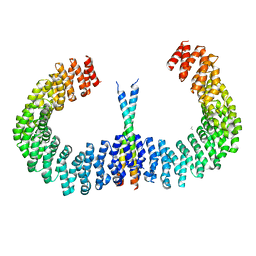 | | A phosphatase complex STRN3-PP2Aa | | Descriptor: | PP2A scaffolding subunit, PROPANE, Striatin-3 | | Authors: | Tang, Y, Zhou, Z.C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Selective Inhibition of STRN3-Containing PP2A Phosphatase Restores Hippo Tumor-Suppressor Activity in Gastric Cancer.
Cancer Cell, 38, 2020
|
|
8XFW
 
 | |
8XFH
 
 | |
6AD1
 
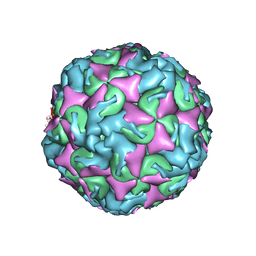 | | The structure of CVA10 procapsid from its complex with Fab 2G8 | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, R, Zheng, Q.B, Xu, L.F, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6AD0
 
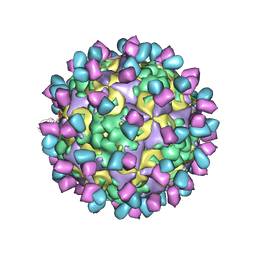 | | The structure of CVA10 mature virion in complex with Fab 2G8 | | Descriptor: | SPHINGOSINE, VH of Fab 2G8, VL of Fab 2G8, ... | | Authors: | Zhu, R, Zheng, Q.B, Xu, L.F, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6ACZ
 
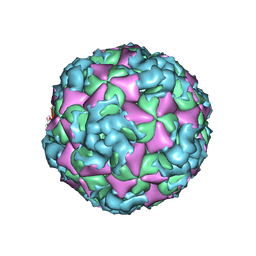 | | The structure of CVA10 virus A-particle from its complex with Fab 2G8 | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, R, Zheng, Q.B, Xu, L.F, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6ACW
 
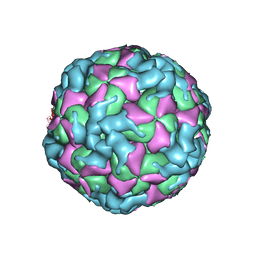 | | The structure of CVA10 virus procapsid particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, R, Xu, L.F, Zheng, Q.B, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6ACY
 
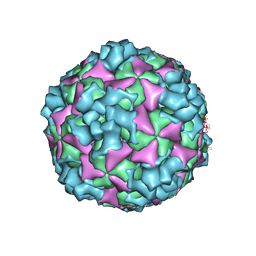 | | The structure of CVA10 virus A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Cui, Y.X, Zheng, Q.B, Zhu, R, Xu, L.F, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
7YSX
 
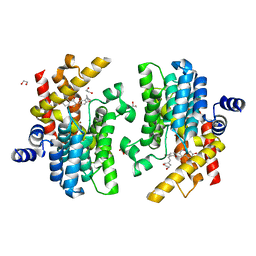 | | Crystal structure of PDE4D complexed with licoisoflavone A | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YQF
 
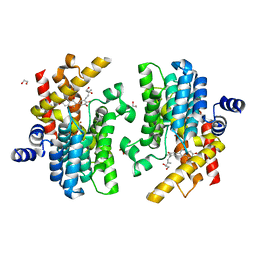 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8XJV
 
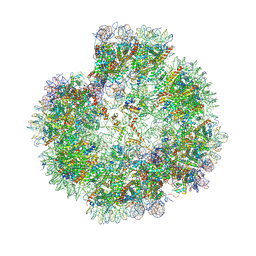 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
1G86
 
 | | CHARCOT-LEYDEN CRYSTAL PROTEIN/N-ETHYLMALEIMIDE COMPLEX | | Descriptor: | CHARCOT-LEYDEN CRYSTAL PROTEIN, N-ETHYLMALEIMIDE | | Authors: | Ackerman, S.J, Liu, L, Kwatia, M.A, Savage, M.P, Leonidas, D.D, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden crystal protein (galectin-10) is not a dual function galectin with lysophospholipase activity but binds a lysophospholipase inhibitor in a novel structural fashion.
J.Biol.Chem., 277, 2002
|
|
7YRD
 
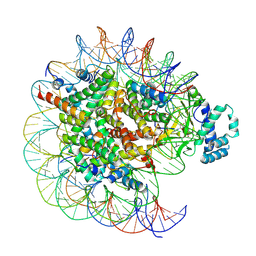 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
7YRG
 
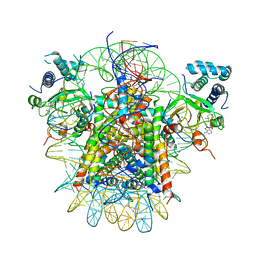 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
6UTK
 
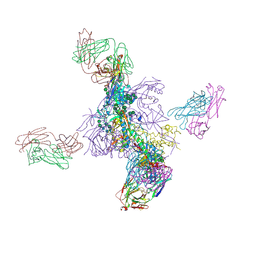 | |
2KBE
 
 | |
5XY9
 
 | | Structure of the MST4 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2017-07-06 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
2KBF
 
 | |
