3LQQ
 
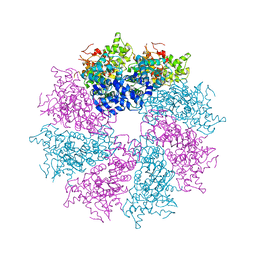 | | Structure of the CED-4 Apoptosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N, Liu, Q. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.534 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
3LQR
 
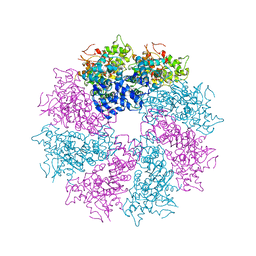 | | Structure of CED-4:CED-3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.896 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
1T29
 
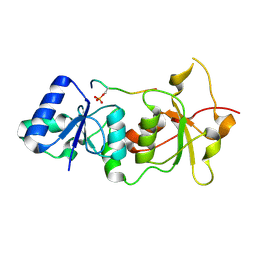 | | Crystal structure of the BRCA1 BRCT repeats bound to a phosphorylated BACH1 peptide | | Descriptor: | BACH1 phosphorylated peptide, Breast cancer type 1 susceptibility protein | | Authors: | Shiozaki, E.N, Gu, L, Yan, N, Shi, Y. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling.
Mol.Cell, 14, 2004
|
|
5XSY
 
 | | Structure of the Nav1.4-beta1 complex from electric eel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein, Voltage-gated sodium channel beta subunit 1, ... | | Authors: | Yan, Z, Zhou, Q, Wu, J.P, Yan, N. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Nav1.4-beta 1 Complex from Electric Eel.
Cell, 170, 2017
|
|
5GJV
 
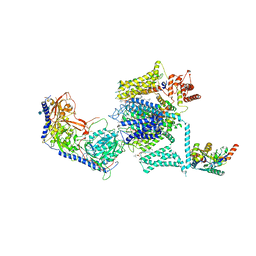 | | Structure of the mammalian voltage-gated calcium channel Cav1.1 complex at near atomic resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Yan, Z, Li, Z.Q, Zhou, Q, Yan, N. | | Deposit date: | 2016-07-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Nature, 537, 2016
|
|
5GJW
 
 | | Structure of the mammalian voltage-gated calcium channel Cav1.1 complex for ClassII map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Yan, Z, Li, Z.Q, Zhou, Q, Yan, N. | | Deposit date: | 2016-07-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Nature, 537, 2016
|
|
5GKZ
 
 | | Structure of RyR1 in a closed state (C3 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
5GL0
 
 | | Structure of RyR1 in a closed state (C4 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
5GL1
 
 | | Structure of RyR1 in an open state | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
5GKY
 
 | | Structure of RyR1 in a closed state (C1 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
8AMX
 
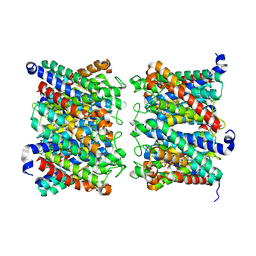 | | AQP7 dimer of tetramers_D4 | | Descriptor: | Aquaporin-7 | | Authors: | Huang, P, Venskutonyte, R, Fan, X, Li, P, Yan, N, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure supports a role of AQP7 as a junction protein.
Nat Commun, 14, 2023
|
|
8AMW
 
 | | AQP7 dimer of tetramers_C1 | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Huang, P, Venskutonyte, R, Fan, X, Li, P, Yan, N, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure supports a role of AQP7 as a junction protein.
Nat Commun, 14, 2023
|
|
8WA2
 
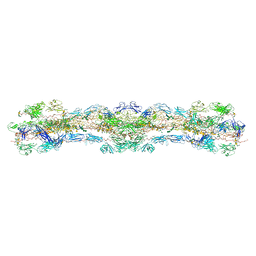 | | cryo-EM structure of native mastigonemes isolated from Chlamydomonas reinhardtii at 3.0 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, J, Tao, H, Chen, J, Pan, J, Yan, C, Yan, N. | | Deposit date: | 2023-09-06 | | Release date: | 2024-04-10 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-guided discovery of protein and glycan components in native mastigonemes.
Cell, 187, 2024
|
|
3JD8
 
 | | cryo-EM structure of the full-length human NPC1 at 4.4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Zhou, Q, Yan, N. | | Deposit date: | 2016-05-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | The structure of Niemann-Pick C1 protein
To Be Published
|
|
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10 | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
3O7P
 
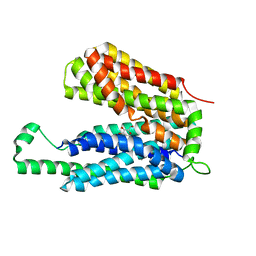 | | Crystal structure of the E.coli Fucose:proton symporter, FucP (N162A) | | Descriptor: | L-fucose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Dang, S.Y, Sun, L.F, Wang, J, Yan, N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of a fucose transporter in an outward-open conformation
Nature, 467, 2010
|
|
4M59
 
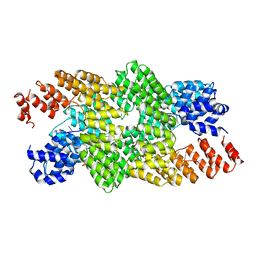 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
4N0G
 
 | | Crystal Structure of PYL13-PP2CA complex | | Descriptor: | Abscisic acid receptor PYL13, MAGNESIUM ION, Protein phosphatase 2C 37, ... | | Authors: | Li, W, Wang, L, Sheng, X, Yan, C, Zhou, R, Hang, J, Yin, P, Yan, N. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13
Cell Res., 23, 2013
|
|
4OIC
 
 | | Crystal structrual of a soluble protein | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Bet v I allergen-like, CHLORIDE ION, ... | | Authors: | He, Y, Hao, Q, Li, W, Yan, C, Yan, N, Yin, P. | | Deposit date: | 2014-01-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Identification and characterization of ABA receptors in Oryza sativa
Plos One, 9, 2014
|
|
4OST
 
 | | Crystal structure of the S505C mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
4OSK
 
 | | Crystal structure of TAL effector reveals the recognition between asparagine and guanine | | Descriptor: | DNA (5'-D(*AP*GP*TP*CP*TP*AP*GP*TP*AP*GP*TP*TP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*AP*AP*CP*TP*AP*CP*TP*AP*GP*AP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
4OT0
 
 | | Crystal structure of the S505T mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
4OSS
 
 | | Crystal structure of the S505Q mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*CP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*GP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
4OSQ
 
 | | Crystal structure of the S505R mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*GP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, ... | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
4OSL
 
 | | Crystal structure of TAL effector reveals the recognition between histidine and guanine | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*CP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*GP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, ... | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
