2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
4AOI
 
 | | Crystal structure of C-MET kinase domain in complex with 4-(3-((1H- pyrrolo(2,3-b)pyridin-3-yl)methyl)-(1,2,4)triazolo(4,3-b)(1,2,4) triazin-6-yl)benzonitrile | | Descriptor: | 4-[3-(1H-pyrrolo[2,3-b]pyridin-3-ylmethyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-6-yl]benzenecarbonitrile, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
1RGI
 
 | | Crystal structure of gelsolin domains G1-G3 bound to actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Burtnick, L.D, Urosev, D, Irobi, E, Narayan, K, Robinson, R.C. | | Deposit date: | 2003-11-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF
Embo J., 23, 2004
|
|
3ZZE
 
 | | Crystal structure of C-MET kinase domain in complex with N'-((3Z)-4- chloro-7-methyl-2-oxo-1,2-dihydro-3H-indol-3-ylidene)-2-(4- hydroxyphenyl)propanohydrazide | | Descriptor: | (2S)-N'-[(3R)-4-chloro-7-methyl-2-oxo-2,3-dihydro-1H-indol-3-yl]-2-(4-hydroxyphenyl)propanehydrazide, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Deng, Y, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-31 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
3ZBX
 
 | | X-ray Structure of c-Met kinase in complex with inhibitor 6-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl) quinoline. | | Descriptor: | 6-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J, Ryan, K. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
3ZC5
 
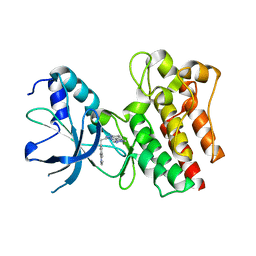 | | X-ray Structure of c-Met kinase in complex with inhibitor (S)-6-(1-(6- (1-methyl-1H-pyrazol-4-yl)-(1,2,4)triazolo(4,3-b)pyridazin-3-yl)ethyl) quinoline. | | Descriptor: | 6-{(1S)-1-[6-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]ethyl}quinoline, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
3ZCL
 
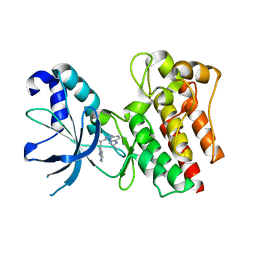 | | X-ray Structure of c-Met kinase in complex with inhibitor (S)-3-(1-(1H-pyrrolo(2,3-b)pyridin-3-yl)ethyl)-N-isopropyl-(1,2,4)triazolo(4,3- b)pyridazin-6-amine | | Descriptor: | (S)-3-(1-(1H-pyrrolo(2,3-b)pyridin-3-yl)ethyl)-N-isopropyl-(1,2,4)triazolo(4,3-b)pyridazin-6-amine, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K. | | Deposit date: | 2012-11-20 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
3ZXZ
 
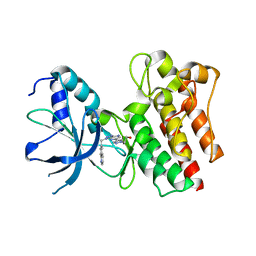 | | X-ray Structure of PF-04217903 bound to the kinase domain of c-Met | | Descriptor: | 2-{4-[1-(QUINOLIN-6-YLMETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRAZIN-6-YL]-1H-PYRAZOL-1-YL}ETHANOL, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-16 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
6XHL
 
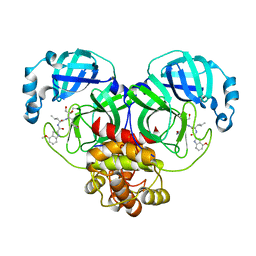 | | Covalent complex of SARS-CoV main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHM
 
 | | Covalent complex of SARS-CoV-2 main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHO
 
 | | Covalent complex of SARS-CoV main protease with ethyl (4R)-4-({N-[(4-methoxy-1H-indol-2-yl)carbonyl]-L-leucyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, ethyl (2E,4S)-4-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHN
 
 | | Covalent complex of SARS-CoV main protease with 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Descriptor: | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate, 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
3VKQ
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with middle X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
2HBX
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
3VKS
 
 | | Assimilatory nitrite reductase (Nii3) - NO complex from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRIC OXIDE, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3VLY
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - SO3 partial complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3VIQ
 
 | | Crystal structure of Swi5-Sfr1 complex from fission yeast | | Descriptor: | GLYCEROL, Mating-type switching protein swi5, NITRATE ION, ... | | Authors: | Kuwabara, N, Murayama, Y, Hashimoto, H, Kokabu, Y, Ikeguchi, M, Sato, M, Mayanagi, K, Tsutsui, Y, Iwasaki, H, Shimizu, T. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Structure, 20, 2012
|
|
3VM0
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - NO2 complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
7Y8Q
 
 | | Amyloid-beta assemblage on GM1-containing membranes | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yagi-Utsumi, M, Itoh, S.G, Okumura, H, Yanagisawa, K, Kato, K, Nishimura, K. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The Double-Layered Structure of Amyloid-beta Assemblage on GM1-Containing Membranes Catalytically Promotes Fibrillization.
Acs Chem Neurosci, 14, 2023
|
|
3VM1
 
 | | assimilatory nitrite reductase (Nii3) - N226K mutant - HCO3 complex from tobacco leaf | | Descriptor: | BICARBONATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
2HBV
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
3VKP
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with low X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3VLX
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - ligand free form from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3VKR
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with high X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3VLZ
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - SO3 full complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
