5EF6
 
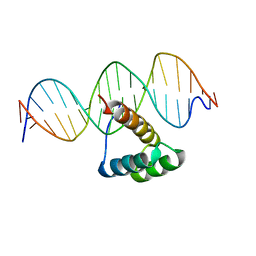 | | Structure of HOXB13 complex with methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
5EHR
 
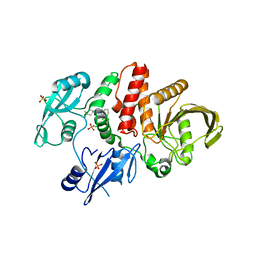 | |
4J7R
 
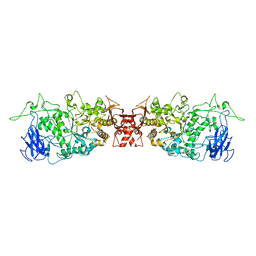 | |
5H6I
 
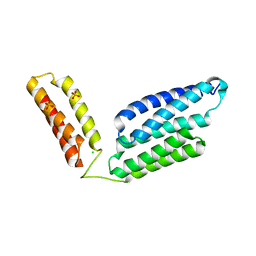 | | Crystal Structure of GBS CAMP Factor | | Descriptor: | CHLORIDE ION, Protein B, SULFATE ION | | Authors: | Jin, T.C, Brefo-Mensah, E.K. | | Deposit date: | 2016-11-13 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of theStreptococcus agalactiaeCAMP factor provides insights into its membrane-permeabilizing activity.
J.Biol.Chem., 293, 2018
|
|
4K2F
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A08522488 | | Descriptor: | (2S)-(4-chlorophenyl)(6-chloropyridin-2-yl)ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
4K2G
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A33442372 | | Descriptor: | (2S)-(4-fluorophenyl)[6-(trifluoromethyl)pyridin-2-yl]ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
8BA4
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BA3
 
 | | Crystal structure of JAK2 JH2 in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B99
 
 | | Crystal structure of JAK2 JH2-V617F in complex with JNJ-7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8U
 
 | |
8B9H
 
 | | Crystal structure of JAK2 JH2 in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B9E
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BAK
 
 | |
8BAB
 
 | | Crystal structure of JAK2 JH2-V617F in complex with CB76 | | Descriptor: | 6-[(1-methylimidazol-2-yl)sulfanylmethyl]-~{N}4-(3-methylphenyl)-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8N
 
 | |
8BA2
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A1 | | Descriptor: | 6-[[(5-bromanylthiophen-2-yl)methyl-methyl-amino]methyl]-~{N}4-(4-methylphenyl)-1,3,5-triazine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8EX1
 
 | |
8EX0
 
 | |
8EX2
 
 | |
5EHP
 
 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP836 | | Descriptor: | 5-[2,3-bis(chloranyl)phenyl]-2-[(3~{R},5~{S})-3,5-dimethylpiperazin-1-yl]pyrimidin-4-amine, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor.
J.Med.Chem., 59, 2016
|
|
1QSU
 
 | | CRYSTAL STRUCTURE OF THE TRIPLE-HELICAL COLLAGEN-LIKE PEPTIDE, (PRO-HYP-GLY)4-GLU-LYS-GLY(PRO-HYP-GLY)5 | | Descriptor: | PROTEIN ((PRO-HYP-GLY)4- GLU-LYS-GLY(PRO-HYP-GLY)5) | | Authors: | Kramer, R.Z, Venugopal, M, Bella, J, Brodsky, B, Berman, H.M. | | Deposit date: | 1999-06-23 | | Release date: | 2000-08-30 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Staggered molecular packing in crystals of a collagen-like peptide with a single charged pair.
J.Mol.Biol., 301, 2000
|
|
5DGG
 
 | | Central domain of uncharacterized Lpg1148 protein from Legionella pneumophila | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Evdokimova, E, Yim, V, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol. Syst. Biol., 12, 2016
|
|
5Y2G
 
 | | Structure of MBP tagged GBS CAMP | | Descriptor: | Maltose-binding periplasmic protein,Protein B, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2017-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4OKD
 
 | |
6C06
 
 | | Mycobacterium tuberculosis RNAP Holo/RbpA/Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
