6JUD
 
 | | Radiation damage in Aspergillus oryzae pro-tyrosinase oxygen-bound C92A/H103F mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JUC
 
 | | Aspergillus oryzae pro-tyrosinase oxygen-bound C92A/H103F mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU5
 
 | | Aspergillus oryzae pro-tyrosinase C92A/F513Y mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5ZBY
 
 | |
8I53
 
 | |
6JU7
 
 | | Aspergillus oryzae active-tyrosinase copper-depleted C92A mutant complexed with L-tyrosine | | Descriptor: | NITRATE ION, TYROSINE, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7F8D
 
 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) G218Y mutant | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | Deposit date: | 2021-07-02 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Increasing loop flexibility affords low-temperature adaptation of a moderate thermophilic malate dehydrogenase from Geobacillus stearothermophilus.
Protein Eng.Des.Sel., 34, 2021
|
|
6JU8
 
 | | Aspergillus oryzae active-tyrosinase copper-bound C92A mutant | | Descriptor: | COPPER (II) ION, NITRATE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JUB
 
 | | Radiation damage in Aspergillus oryzae pro-tyrosinase oxygen-bound C92A mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3W6V
 
 | | Crystal structure of the DNA-binding domain of AdpA, the global transcriptional factor, in complex with a target DNA | | Descriptor: | AdpA, DNA (5'-D(*AP*GP*GP*TP*TP*GP*GP*CP*GP*GP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*TP*GP*TP*GP*AP*AP*CP*CP*CP*GP*CP*CP*AP*AP*C)-3') | | Authors: | Yao, M.D, Ohtsuka, J, Nagata, K, Miyazono, K, Ohnishi, Y, Tanokura, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Complex Structure of the DNA-binding Domain of AdpA, the Global Transcription Factor in Streptomyces griseus, and a Target Duplex DNA Reveals the Structural Basis of Its Tolerant DNA Sequence Specificity
J.Biol.Chem., 288, 2013
|
|
6JU4
 
 | | Aspergillus oryzae pro-tyrosinase F513Y mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JUA
 
 | | Aspergillus oryzae pro-tyrosinase oxygen-bound C92A mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3VS9
 
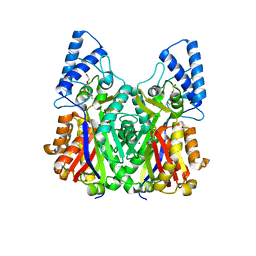 | | Crystal structure of type III PKS ArsC mutant | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
6JU9
 
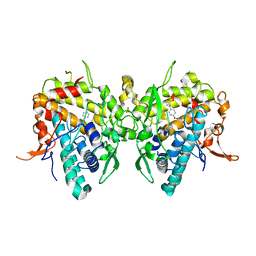 | | Aspergillus oryzae active-tyrosinase copper-bound C92A mutant complexed with L-tyrosine | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, COPPER (II) ION, NITRATE ION, ... | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7DTI
 
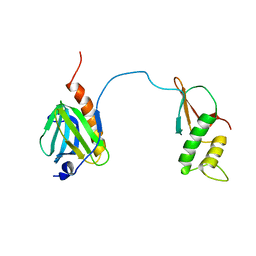 | |
7DTH
 
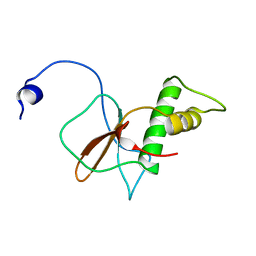 | | Solution structure of RPB6, common subunit of RNA polymerases I, II, and III | | Descriptor: | DNA-directed RNA polymerases I, II, and III subunit RPABC2 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three human RNA polymerases interact with TFIIH via a common RPB6 subunit.
Nucleic Acids Res., 50, 2022
|
|
7X8V
 
 | | Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity | | Descriptor: | Os01g0156300 protein | | Authors: | Ichimaru, K, Harada, K, Yamaguchi, K, Shigeta, S, Shimada, K, Ishikawa, K, Inoue, K, Nishio, Y, Yoshimura, S, Inoue, H, Yamashita, E, Fujiwara, T, Nakagawa, A, Kojima, C, Kawasaki, T. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity.
Nat Commun, 13, 2022
|
|
3VGK
 
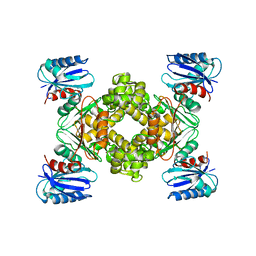 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus | | Descriptor: | Glucokinase, SULFATE ION, ZINC ION | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
3VM6
 
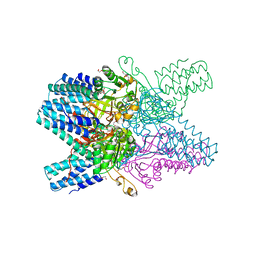 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1 in complex with alpha-D-ribose-1,5-bisphosphate | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, A, Fujihashi, M, Aono, R, Sato, T, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2011-12-08 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
5YY0
 
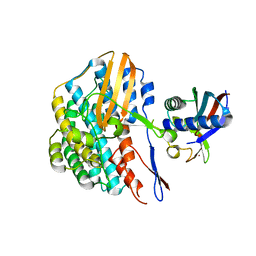 | | Crystal structure of the HyhL-HypA complex (form II) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.243 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YXY
 
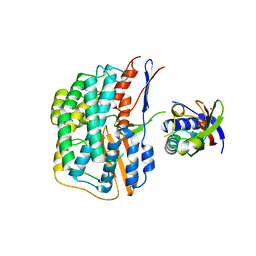 | | Crystal structure of the HyhL-HypA complex (form I) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2DKV
 
 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, chitinase | | Authors: | Kezuka, Y, Nishizawa, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
6K5W
 
 | | Solution structure of the chromodomain of yeast Eaf3 | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eaf3 chromodomain acts as a pH sensor for gene expression by altering its binding affinity for histone methylated-lysine residues.
Biosci.Rep., 40, 2020
|
|
3VXF
 
 | | X/N Joint refinement of Human alpha-thrombin-Bivalirudin complex PD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.602 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
3VGM
 
 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose | | Descriptor: | Glucokinase, POTASSIUM ION, ZINC ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
