3U2R
 
 | |
3U2E
 
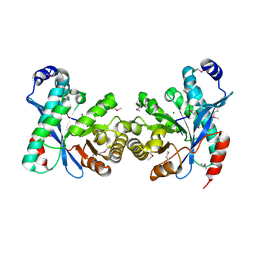 | | EAL domain of phosphodiesterase PdeA in complex with 5'-pGpG and Mg++ | | Descriptor: | GGDEF family protein, MAGNESIUM ION, RNA (5'-R(P*GP*G)-3') | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | EAL domain from Caulobacter crescentus in complex with 5'-pGpG and Mg++
To be Published
|
|
2NP3
 
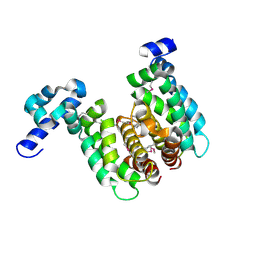 | | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3. | | Descriptor: | Putative TetR-family regulator | | Authors: | Koclega, K.D, Xu, X, Chruszcz, M, Gu, J, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3.
To be Published
|
|
2NP5
 
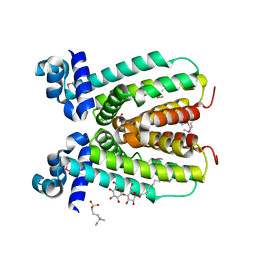 | | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Kagan, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1.
TO BE PUBLISHED
|
|
3LV9
 
 | |
3NRV
 
 | |
3MAJ
 
 | | Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris CGA009 | | Descriptor: | DNA processing chain A, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris
To be Published
|
|
3FET
 
 | | Crystal Structure of the Electron Transfer Flavoprotein Subunit Alpha related Protein Ta0212 from Thermoplasma acidophilum | | Descriptor: | Electron transfer flavoprotein subunit alpha related protein | | Authors: | Kim, Y, Tesar, C, Souvong, K, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-01 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Electron Transfer Flavoprotein Subunit Alpha related Protein Ta0212 from Thermoplasma acidophilum
To be Published
|
|
3NRT
 
 | | The crystal structure of putative ryanodine receptor from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative ryanodine receptor | | Authors: | Wu, R, Zhang, R, James, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | he crystal strucutre of putative ryanodine receptor from Bacteroides thetaiotaomicron VPI-5482
To be Published
|
|
3OVK
 
 | |
3OLO
 
 | | Crystal structure of a PAS domain from two-component sensor histidine kinase | | Descriptor: | GLYCEROL, Two-component sensor histidine kinase | | Authors: | Michalska, K, Chhor, G, Bearden, J, Fenske, R.J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal structure of a PAS domain from two-component sensor histidine kinase
To be Published
|
|
3OJ0
 
 | | Crystal structure of glutamyl-tRNA reductase from Thermoplasma volcanium (nucleotide binding domain) | | Descriptor: | GLYCEROL, Glutamyl-tRNA reductase, SULFATE ION | | Authors: | Michalska, K, Marshall, N, Clancy, S, Puttagunta, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Crystal structure of glutamyl-tRNA reductase from Thermoplasma volcanium (nucleotide binding domain)
To be Published
|
|
6DZG
 
 | |
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4W9R
 
 | | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271
To Be Published
|
|
3UKJ
 
 | | Crystal structure of extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, GLYCEROL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-09 | | Release date: | 2011-11-23 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
6HCD
 
 | | Structure of universal stress protein from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | Authors: | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-08-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
4OPF
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS8 | | Descriptor: | NRPS/PKS | | Authors: | Osipiuk, J, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1VPD
 
 | | X-Ray Crystal Structure of Tartronate Semialdehyde Reductase [Salmonella Typhimurium LT2] | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, TARTRONATE SEMIALDEHYDE REDUCTASE | | Authors: | Osipiuk, J, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-22 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
J Struct Funct Genomics, 10, 2009
|
|
1VZY
 
 | | Crystal structure of the Bacillus subtilis HSP33 | | Descriptor: | 33 KDA CHAPERONIN, ACETATE ION, ZINC ION | | Authors: | Janda, I.K, Devedjiev, Y, Derewenda, U, Dauter, Z, Bielnicki, J, Cooper, D.R, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-29 | | Release date: | 2004-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism.
Structure, 12, 2004
|
|
6DM4
 
 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila in complex with Homo sapiens Shc1 phospho-Tyr317 peptide | | Descriptor: | RavO, SULFATE ION, Shc1 phospho-Tyr317 peptide | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila in complex with Homo sapiens Shc1 phospho-Tyr317 peptide
To Be Published
|
|
6DM3
 
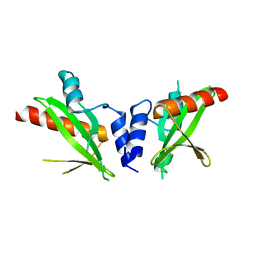 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein | | Descriptor: | RavO | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein
To Be Published
|
|
4W97
 
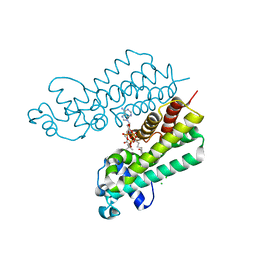 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4W66
 
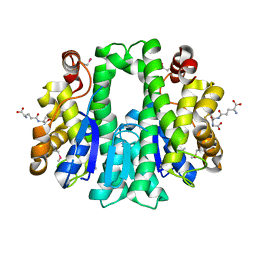 | |
4EYO
 
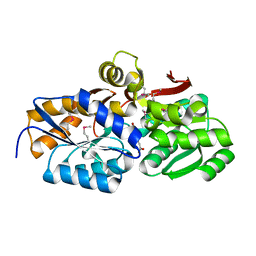 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
