1M42
 
 | |
1OQ6
 
 | | solution structure of Copper-S46V CopA from Bacillus subtilis | | Descriptor: | COPPER (II) ION, Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, l, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
1P6T
 
 | | Structure characterization of the water soluble region of P-type ATPase CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the function of the N-terminal domain of the ATPase CopA from Bacillus subtilis.
J.Biol.Chem., 278, 2003
|
|
1P8G
 
 | |
1S6O
 
 | | Solution structure and backbone dynamics of the apo-form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-26 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1ON4
 
 | | Solution structure of soluble domain of Sco1 from Bacillus Subtilis | | Descriptor: | Sco1 | | Authors: | Balatri, E, Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-02-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Sco1: A Thioredoxin-like Protein Involved in Cytochrome c Oxidase Assembly
STRUCTURE, 11, 2003
|
|
1OPZ
 
 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonneli, L, Su, X.C. | | Deposit date: | 2003-03-06 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis.
J.Mol.Biol., 331, 2003
|
|
1PFD
 
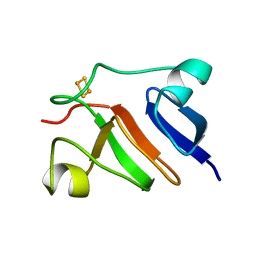 | | THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN, NMR, 18 STRUCTURES | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Im, S.-C, Liu, G, Luchinat, C, Sykes, A.G, Bertini, I. | | Deposit date: | 1998-05-05 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of parsley [2Fe-2S]ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
2JT5
 
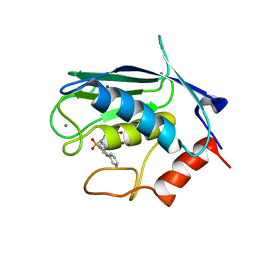 | | solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of n-hydroxy-2-[n-(2-hydroxyethyl)biphenyl-4-sulfonamide] hydroxamic acid (MLC88) | | Descriptor: | CALCIUM ION, N~2~-(biphenyl-4-ylsulfonyl)-N-hydroxy-N~2~-(2-hydroxyethyl)glycinamide, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JNP
 
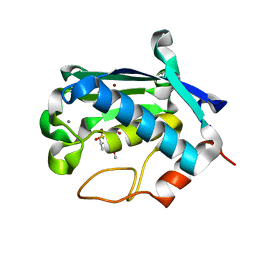 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-3, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JT6
 
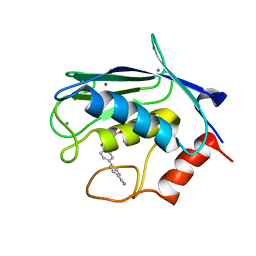 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of 3-4'-cyanobyphenyl-4-yloxy)-n-hdydroxypropionamide (MMP-3 inhibitor VII) | | Descriptor: | 3-[(4'-cyanobiphenyl-4-yl)oxy]-N-hydroxypropanamide, CALCIUM ION, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-23 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2K70
 
 | | Solution structures of copper loaded form PCuA (cis conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P.A, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K6V
 
 | | Solution structures of apo Sco1 protein from Thermus Thermophilus | | Descriptor: | Putative cytochrome c oxidase assembly protein | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K9C
 
 | | Paramagnetic shifts in solid-state NMR of Proteins to elicit structural information | | Descriptor: | COBALT (II) ION, Macrophage metalloelastase | | Authors: | Balayssac, S, Bertini, I, Bhaumik, A, Lelli, M, Luchinat, C. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic shifts in solid-state NMR of proteins to elicit structural information
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2K6W
 
 | | Solution structures of apo PCuA (trans conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K6Y
 
 | | Solution structures of apo form PCuA (cis conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2KMX
 
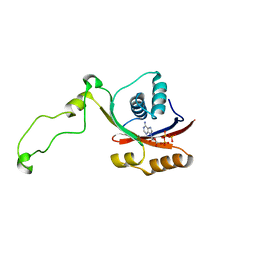 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
2LDI
 
 | |
2L50
 
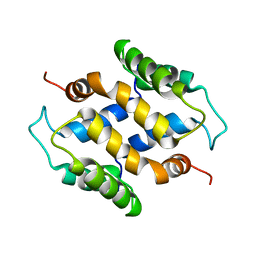 | | Solution structure of apo S100A16 | | Descriptor: | Protein S100-A16 | | Authors: | Babini, E, Bertini, I, Borsi, V, Calderone, V, Hu, X, Luchinat, C, Parigi, G. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of human S100A16, a low-affinity calcium binder.
J.Biol.Inorg.Chem., 16, 2011
|
|
2LU5
 
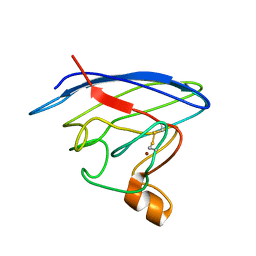 | | Structure and chemical shifts of Cu(I),Zn(II) superoxide dismutase by solid-state NMR | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn] | | Authors: | Knight, M.J, Pell, A.J, Bertini, I, Felli, I.C, Gonnelli, L, Pierattelli, R, Herrmann, T, Emsley, L, Pintacuda, G. | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2JQA
 
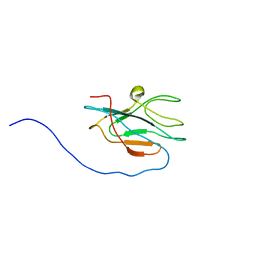 | | Solution structure of apo-DR1885 from Deinococcus radiodurans | | Descriptor: | Hypothetical protein | | Authors: | Banci, L, Bertini, I, Ciofi Baffoni, S, Katsari, E, Katsaros, N, Kubicek, K. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2JSC
 
 | | NMR structure of the cadmium metal-sensor CMTR from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Transcriptional regulator Rv1994c/MT2050 | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Cavet, J.S, Dennison, C, Graham, A.I, Harvie, D.R, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-02 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Analysis of Cadmium Sensing by Winged Helix Repressor CmtR.
J.Biol.Chem., 282, 2007
|
|
2K6Z
 
 | | Solution structures of copper loaded form PCuA (trans conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P.A, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2L4D
 
 | | cytochrome c domain of pp3183 protein from Pseudomonas putida | | Descriptor: | HEME C, SCO1/SenC family protein/cytochrome c | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Kozyreva, T, Mori, M, Wang, S. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Sco proteins are involved in electron transfer processes
J.Biol.Inorg.Chem., 16, 2011
|
|
2JSD
 
 | | Solution structure of MMP20 complexed with NNGH | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-20, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Arendt, Y, Banci, L, Bertini, I, Cantini, F, Cozzi, R, Del Conte, R, Gonnelli, L, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-03 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Catalytic domain of MMP20 (Enamelysin) - the NMR structure of a new matrix metalloproteinase.
Febs Lett., 581, 2007
|
|
