4FAF
 
 | | Substrate CA/p2 in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, substrate CA/p2 peptide | | Authors: | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
4FAE
 
 | | Substrate p2/NC in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, Substrate p2/NC peptide | | Authors: | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
5H6Y
 
 | |
4JFI
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione | | Descriptor: | 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4K2C
 
 | | HSA Ligand Free | | Descriptor: | Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Huang, M. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin.
J. Biol. Chem., 288, 2013
|
|
4IW2
 
 | | HSA-glucose complex | | Descriptor: | D-glucose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Luo, Z, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
4JFK
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFL
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one | | Descriptor: | 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFM
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate | | Descriptor: | 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
8AOU
 
 | |
9IVD
 
 | |
7KRB
 
 | |
4IW1
 
 | | HSA-fructose complex | | Descriptor: | D-fructose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
4JFJ
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
5UHR
 
 | |
7XEI
 
 | | SARS-CoV-2-prototyped-RBD and CB6-092-Fab complex | | Descriptor: | CB6-092-Fab heavy chain, CB6-092-Fab light chain, Spike protein S1 | | Authors: | Wang, Y, Feng, Y. | | Deposit date: | 2022-03-31 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | SARS-CoV-2-prototyped-RBD and CB6-092-Fab complex
To Be Published
|
|
7XEG
 
 | | SARS-CoV-2-Beta-RBD and CB6-092-Fab complex | | Descriptor: | CB6-092-Fab heavy chain, CB6-092-Fab light chain, Spike protein S1 | | Authors: | Wang, Y, Feng, Y. | | Deposit date: | 2022-03-31 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | SARS-CoV-2-Beta-RBD and CB6-092-Fab complex
To Be Published
|
|
5UJT
 
 | | Crystal structure of human HLA-DQ8 in complex with insulin mimotope binding in register 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DQ-beta-1, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8XIQ
 
 | | Structure of L796778-SSTR3 G protein complex | | Descriptor: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XIO
 
 | | Structure of L797591-SSTR1 G protein complex | | Descriptor: | (2~{S})-~{N}-[(4~{S})-6-azanyl-2,2,4-trimethyl-hexyl]-3-naphthalen-1-yl-2-[[2-phenylethyl(2-pyridin-2-ylethyl)carbamoyl]amino]propanamide, G-alpha-i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XIR
 
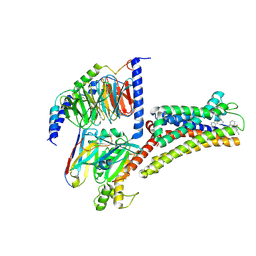 | | Structure of pasireotide-SSTR3 G protein complex | | Descriptor: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XIP
 
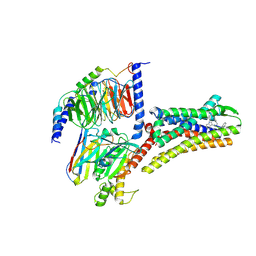 | | Structure of Pasireotide-SSTR1 G protein complex | | Descriptor: | 004-DTR-LYS-TY5-PHE-A1D5E, G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5WYI
 
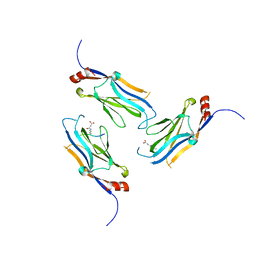 | | The Yaf9 YEATS domain Recognizing H3K122suc Peptide | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, ILE-MET-PRO-LYS-ASP-ILE-GLN-LEU, SUCCINIC ACID, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-17 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Yaf9 YEATS domain Recognizing H3K122suc Peptide
To Be Published
|
|
5W87
 
 | |
8VV0
 
 | |
