1J9B
 
 | | ARSENATE REDUCTASE+0.4M ARSENITE FROM E. COLI | | Descriptor: | ARSENATE REDUCTASE, CESIUM ION, SULFATE ION, ... | | Authors: | Martin, P, Edwards, B.F. | | Deposit date: | 2001-05-24 | | Release date: | 2001-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme.
Structure, 9, 2001
|
|
4P6X
 
 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
4P6W
 
 | | Crystal Structure of mometasone furoate-bound glucocorticoid receptor ligand binding domain | | Descriptor: | Glucocorticoid receptor, MOMETASONE FUROATE, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
3R2F
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with BMS-693391 AKA (2S)-2-((3R)-3-acetamido-3-isobutyl-2-oxo-1-pyrrolidinyl)-N-((1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-2-((2R,4R)-4-propoxy-2-pyrrolidinyl)ethyl)-4-phenylbutanamide | | Descriptor: | (2S)-2-[(3R)-3-(acetylamino)-3-(2-methylpropyl)-2-oxopyrrolidin-1-yl]-N-{(1R,2S)-3-(3,5-difluorophenyl)-1-hydroxy-1-[(2R,4R)-4-propoxypyrrolidin-2-yl]propan-2-yl}-4-phenylbutanamide, Beta-secretase 1 | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2011-03-14 | | Release date: | 2011-08-31 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Monosubstituted {gamma}-lactam and conformationally constrained 1,3-diaminopropan-2-ol transition-state isostere inhibitors of {beta}-secretase (BACE).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SKG
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with (2S)-2-((3R)-3-acetamido-3-isobutyl-2-oxo-1-pyrrolidinyl)-N-((1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-2-(1,2,3,4-tetrahydro-3-isoquinolinyl)ethyl)-4-phenylbutanamide | | Descriptor: | (2S)-2-[(3R)-3-(acetylamino)-3-(2-methylpropyl)-2-oxopyrrolidin-1-yl]-N-{(1R,2S)-3-(3,5-difluorophenyl)-1-hydroxy-1-[(3R)-1,2,3,4-tetrahydroisoquinolin-3-yl]propan-2-yl}-4-phenylbutanamide, Beta-secretase 1 | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2011-06-22 | | Release date: | 2011-09-07 | | Last modified: | 2012-12-05 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Synthesis and in vivo evaluation of cyclic diaminopropane BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SHZ
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-chloro-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SHY
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-fluoro-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SIE
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SKF
 
 | |
5W5O
 
 | |
5W5J
 
 | |
6A5H
 
 | |
6ATV
 
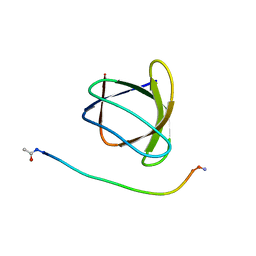 | | The molecular mechanisms by which NS1 of the 1918 Spanish influenza A virus hijack host protein-protein interactions | | Descriptor: | Adapter molecule crk, proline-rich motif in IAV-NS1 | | Authors: | Shen, Q, Zeng, D, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Molecular Mechanisms of Tight Binding through Fuzzy Interactions.
Biophys. J., 114, 2018
|
|
5XX9
 
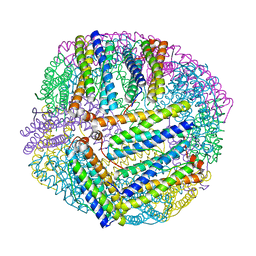 | |
6LJJ
 
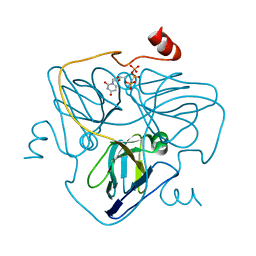 | | Swine dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 1, ... | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
5XTZ
 
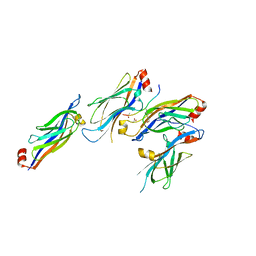 | | Crystal structure of GAS41 YEATS bound to H3K27ac peptide | | Descriptor: | ACETATE ION, THR-LYS-ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, YEATS domain-containing protein 4 | | Authors: | Li, H.T, Zhao, D. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity.
Cell Discov, 4, 2018
|
|
6LIS
 
 | | ASFV dUTPase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LJ3
 
 | |
6LJO
 
 | | African swine fever virus dUTPase | | Descriptor: | E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
1S3D
 
 | | ARSENATE REDUCTASE R60A MUTANT FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | Demel, S, Edwards, B.F. | | Deposit date: | 2004-01-13 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
7Y6G
 
 | |
7Y6P
 
 | | Cryo-EM structure if bacterioferritin holoform | | Descriptor: | Bacterioferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Bacterioferritin nanocage structures uncover the biomineralization process in ferritins.
Pnas Nexus, 2, 2023
|
|
1S3C
 
 | | ARSENATE REDUCTASE C12S MUTANT FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | DeMel, S, Edwards, B.F. | | Deposit date: | 2004-01-13 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
7Y6F
 
 | | Cryo-EM structure of Apo form of ScBfr | | Descriptor: | Bacterioferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Bacterioferritin nanocage structures uncover the biomineralization process in ferritins.
Pnas Nexus, 2, 2023
|
|
1SD8
 
 | | ARSENATE REDUCTASE R60K MUTANT FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | DeMel, S, Edwards, B.F. | | Deposit date: | 2004-02-13 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
