1H37
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, {4-[((1S,2S)-2-{[ALLYL(CYCLOPROPYL)AMINO]METHYL}CYCLOPROPYL)METHOXY]PHENYL}(4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3N
 
 | | Leucyl-tRNA synthetase from Thermus thermophilus complexed with a sulphamoyl analogue of leucyl-adenylate | | Descriptor: | LEUCINE, LEUCYL-TRNA SYNTHETASE, SULFATE ION, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2002-09-11 | | Release date: | 2002-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2A Structure of Leucyl-tRNA Synthetase and its Complex with a Leucyl-Adenylate Analogue
Embo J., 19, 2000
|
|
7QRF
 
 | | Structure of the dimeric complex between precursor membrane ectodomain (prM) and envelope protein ectodomain (E) from tick-borne encephalitis virus | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Envelope protein E, ... | | Authors: | Vaney, M.C, Rouvinski, A, Rey, F.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Nat Commun, 13, 2022
|
|
8Q6L
 
 | | human Carbonic Anhydrase I in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, Carbonic anhydrase 1, SODIUM ION, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-08-13 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | human Carbonic Anhydrase I in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol
To Be Published
|
|
1H0T
 
 | | An affibody in complex with a target protein: structure and coupled folding | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN A, ZSPA-1 AFFIBODY | | Authors: | Wahlberg, E, Lendel, C, Helgstrand, M, Allard, P, Dincbas-Renqvist, V, Hedqvist, A, Berglund, H, Nygren, P.-A, Hard, T. | | Deposit date: | 2002-06-27 | | Release date: | 2003-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Affibody in Complex with a Target Protein: Structure and Coupled Folding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8QXZ
 
 | | Xylanase from Bacillus circulans mutant E78Q/Y69A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX
Febs J., 2024
|
|
8QY2
 
 | | Xylanase from Bacillus circulans mutant E78Q/F125A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX
Febs J., 2024
|
|
8R85
 
 | |
8B1N
 
 | | Crystal structure of TrmD-Tm1570 from Calditerrivibrio nitroreducens in complex with S-adenosyl-L-methionine | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kluza, A, Lewandowska, I, Augustyniak, R, Sulkowska, J. | | Deposit date: | 2022-09-10 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Are there double knots in proteins? Prediction and in vitro verification based on TrmD-Tm1570 fusion from C. nitroreducens.
Front Mol Biosci, 10, 2023
|
|
8QXY
 
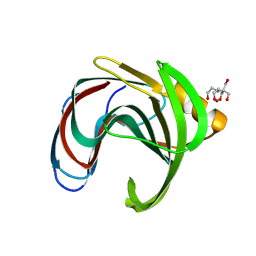 | | Xylanase from Bacillus circulans mutant E78Q bound to xylotriose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX
Febs J., 2024
|
|
8QY0
 
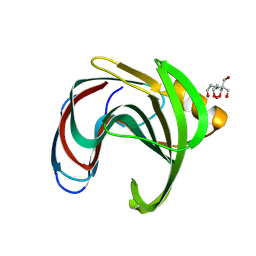 | | Xylanase from Bacillus circulans mutant E78Q/Y69A bound to xylotriose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase, ZINC ION, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX
Febs J., 2024
|
|
8QY1
 
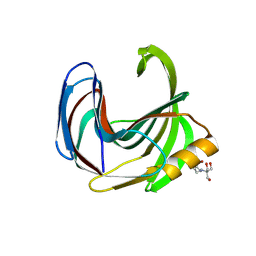 | | Xylanase from Bacillus circulans mutant E78Q/Y69A bound to xylohexaose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX
Febs J., 2024
|
|
8QY3
 
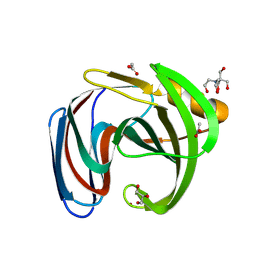 | | Xylanase from Bacillus circulans mutant E78Q/F125A bound to xylotriose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX
Febs J., 2024
|
|
3L92
 
 | | Phosphopantetheine adenylyltransferase from Yersinia pestis complexed with coenzyme A. | | Descriptor: | COENZYME A, Phosphopantetheine adenylyltransferase | | Authors: | Osipiuk, J, Maltseva, N, Makowska-grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray crystal structure of phosphopantetheine adenylyltransferase from Yersinia pestis.
To be Published
|
|
8GAC
 
 | |
8GAD
 
 | |
8GAB
 
 | |
1C3X
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH 8-IODO-GUANINE | | Descriptor: | 8-IODO-GUANINE, CALCIUM ION, PENTOSYLTRANSFERASE, ... | | Authors: | Tebbe, J, Bzowska, A, Wielgus-Kutrowska, B, Schroeder, W, Kazimierczuk, Z, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 1999-07-29 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the purine nucleoside phosphorylase (PNP) from Cellulomonas sp. and its implication for the mechanism of trimeric PNPs.
J.Mol.Biol., 294, 1999
|
|
5T6L
 
 | | Crystal structure of 10E8 Fab in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Antibody 10E8 FAB HEAVY CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
1BUI
 
 | | Structure of the ternary microplasmin-staphylokinase-microplasmin complex: a proteinase-cofactor-substrate complex in action | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, Plasminogen, Staphylokinase | | Authors: | Parry, M.A.A, Fernandez-Catalan, C, Bergner, A, Huber, R, Hopfner, K, Schlott, B, Guehrs, K, Bode, W. | | Deposit date: | 1998-09-04 | | Release date: | 1999-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The ternary microplasmin-staphylokinase-microplasmin complex is a proteinase-cofactor-substrate complex in action.
Nat.Struct.Biol., 5, 1998
|
|
1C0Y
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
5SY8
 
 | |
5T5B
 
 | |
1BZ0
 
 | | HEMOGLOBIN A (HUMAN, DEOXY, HIGH SALT) | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1998-11-04 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Accommodation of insertions in helices: the mutation in hemoglobin Catonsville (Pro 37 alpha-Glu-Thr 38 alpha) generates a 3(10)-->alpha bulge.
Biochemistry, 32, 1993
|
|
3LAD
 
 | |
