2A4Z
 
 | | Crystal Structure of human PI3Kgamma complexed with AS604850 | | Descriptor: | (5E)-5-[(2,2-DIFLUORO-1,3-BENZODIOXOL-5-YL)METHYLENE]-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-06-30 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
4RWA
 
 | | Synchrotron structure of the human delta opioid receptor in complex with a bifunctional peptide (PSI community target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Soluble cytochrome b562,Delta-type opioid receptor, bifunctional peptide | | Authors: | Fenalti, G, Zatsepin, N.A, Betti, C, Giguere, P, Han, G.W, Ishchenko, A, Liu, W, Guillemyn, K, Zhang, H, James, D, Wang, D, Weierstall, U, Spence, J.C.H, Boutet, S, Messerschmidt, M, Williams, G.J, Gati, C, Yefanov, O.M, White, T.A, Oberthuer, D, Metz, M, Yoon, C.H, Barty, A, Chapman, H.N, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Fromme, P, Tourwe, D, Schiller, P.W, Roth, B.L, Ballet, S, Katritch, V, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-12-01 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for bifunctional peptide recognition at human delta-opioid receptor.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3NEW
 
 | | p38-alpha complexed with Compound 10 | | Descriptor: | 4-(trifluoromethyl)-3-[3-(trifluoromethyl)phenyl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Goedken, E.R, Comess, K.M, Sun, C, Argiriadi, M, Jia, Y, Quinn, C.M, Banach, D.L, Marcotte, D, Borhani, D. | | Deposit date: | 2010-06-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases.
Acs Chem.Biol., 6, 2011
|
|
1N50
 
 | |
2A5U
 
 | | Crystal Structure of human PI3Kgamma complexed with AS605240 | | Descriptor: | (5E)-5-(QUINOXALIN-6-YLMETHYLENE)-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
4RWD
 
 | | XFEL structure of the human delta opioid receptor in complex with a bifunctional peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, SODIUM ION, ... | | Authors: | Fenalti, G, Zatsepin, N.A, Betti, C, Giguere, P, Han, G.W, Ishchenko, A, Liu, W, Guillemyn, K, Zhang, H, James, D, Wang, D, Weierstall, U, Spence, J.C.H, Boutet, S, Messerschmidt, M, Williams, G.J, Gati, C, Yefanov, O.M, White, T.A, Oberthuer, D, Metz, M, Yoon, C.H, Barty, A, Chapman, H.N, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Fromme, P, Tourwe, D, Schiller, P.W, Roth, B.L, Ballet, S, Katritch, V, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-12-02 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for bifunctional peptide recognition at human delta-opioid receptor.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1NCS
 
 | | NMR STUDY OF SWI5 ZINC FINGER DOMAIN 1 | | Descriptor: | TRANSCRIPTIONAL FACTOR SWI5, ZINC ION | | Authors: | Dutnall, R.N, Neuhaus, D, Rhodes, D. | | Deposit date: | 1996-02-26 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first zinc finger domain of SWI5: a novel structural extension to a common fold.
Structure, 4, 1996
|
|
1N90
 
 | |
2DAB
 
 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | D-AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
2DAA
 
 | |
4WRT
 
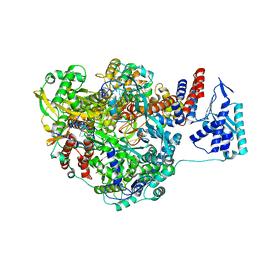 | | Crystal structure of Influenza B polymerase with bound vRNA promoter (form FluB2) | | Descriptor: | Influenza virus polymerase vRNA promoter 3' end, Influenza virus polymerase vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
2CJB
 
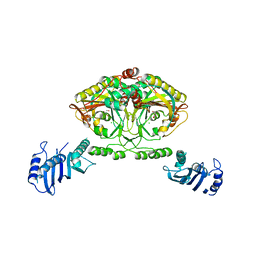 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with serine | | Descriptor: | CHLORIDE ION, SERINE, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-30 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
8ILS
 
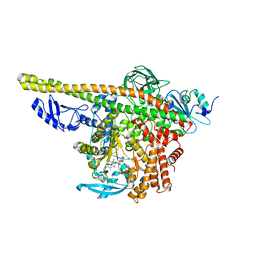 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7PTF
 
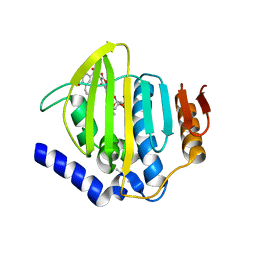 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with novobiocin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B, ... | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PTG
 
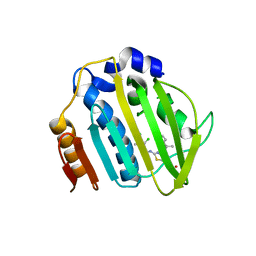 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQI
 
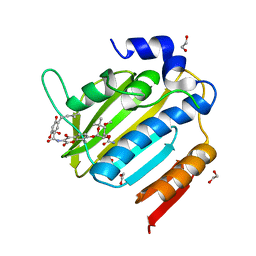 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQM
 
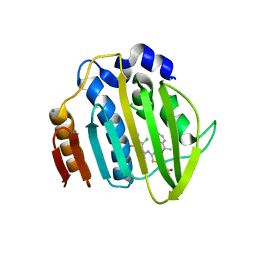 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQL
 
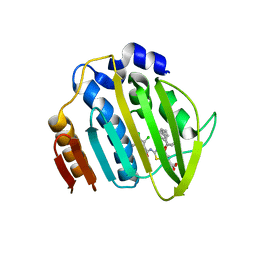 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2704 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1R)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2W
 
 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2M
 
 | | E.coli GyrB24 with inhibitor LMD43 (EBL2560) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
2DNJ
 
 | |
4XDS
 
 | | Deoxyguanosinetriphosphate Triphosphohydrolase from Escherichia coli with Nickel | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, NICKEL (II) ION, SULFATE ION | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
9GAO
 
 | | Crystal structure of CRBNmidi in complex with 2-(4-(2,6-dioxopiperidin-3-yl)phenoxy)-N-methylacetamide | | Descriptor: | 2-[4-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]phenoxy]-~{N}-methyl-ethanamide, Protein cereblon, ZINC ION | | Authors: | Rutter, Z.J, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
