7CJI
 
 | | Photosystem II structure in the S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
5H35
 
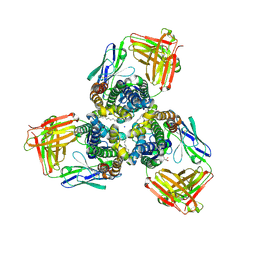 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Sulfolobus solfataricus | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
7CJJ
 
 | | Photosystem II structure in the S2 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7COU
 
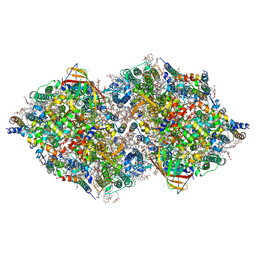 | | Structure of cyanobacterial photosystem II in the dark S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
6IU3
 
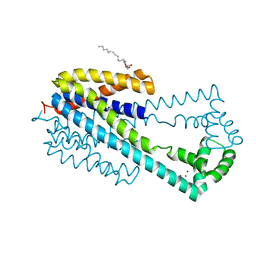 | | Crystal structure of iron transporter VIT1 with zinc ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU9
 
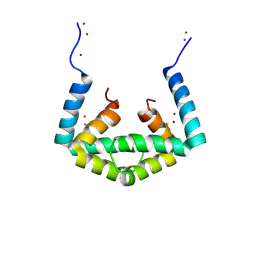 | | Crystal structure of cytoplasmic metal binding domain with iron ions | | Descriptor: | FE (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
6IU5
 
 | | Crystal structure of cytoplasmic metal binding domain with zinc ions | | Descriptor: | CHLORIDE ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU4
 
 | | Crystal structure of iron transporter VIT1 with cobalt ion | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
5YFI
 
 | | Crystal structure of the anti-human prostaglandin E receptor EP4 antibody Fab fragment | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, ZINC ION | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YHL
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and an antagonist Br-derivative | | Descriptor: | 4-[2-[[(2R)-2-(4-bromanylnaphthalen-1-yl)propanoyl]amino]-4-cyano-phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YWY
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | Descriptor: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5Y5F
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5H
 
 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5G
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
6ZHX
 
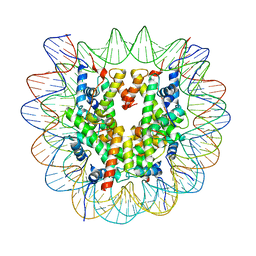 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (145-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Croll, T.I, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
5GIJ
 
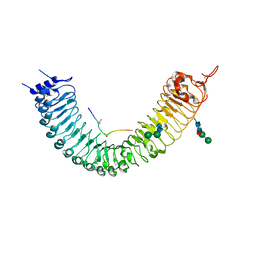 | | Crystal structure of TDR-TDIF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR, ... | | Authors: | Morita, J, Kato, K, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Nat Commun, 7, 2016
|
|
8OOR
 
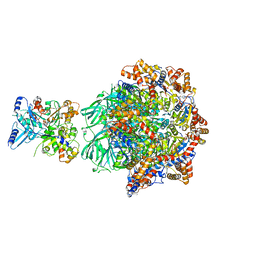 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
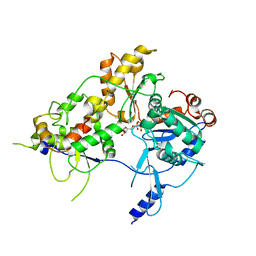 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOP
 
 | | CryoEM Structure INO80core Hexasome complex composite model state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOC
 
 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO7
 
 | | CryoEM Structure INO80core Hexasome complex composite model state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
6BAK
 
 | | The structure of the Stigmatella aurantiaca phytochrome chromophore binding domain T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-13 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAO
 
 | | Stigmatella aurantiaca phytochrome photosensory core module, wild type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
