5OPG
 
 | | Structure of the Hantaan virus Gn glycoprotein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Rissanen, I, Stass, R, Zeltina, A, Li, S, Hepojoki, J, Harlos, K, Gilbert, R.J.C, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2017-08-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Transitions of the Conserved and Metastable Hantaviral Glycoprotein Envelope.
J. Virol., 91, 2017
|
|
1SPX
 
 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
1SCZ
 
 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
1T9F
 
 | | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function | | Descriptor: | MALONATE ION, protein 1d10 | | Authors: | Symersky, J, Li, S, Bunzel, R, Schormann, N, Luo, D, Huang, W.Y, Qiu, S, Gray, R, Zhang, Y, Arabashi, A, Lu, S, Luan, C.H, Tsao, J, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-16 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function.
To be Published
|
|
6VQV
 
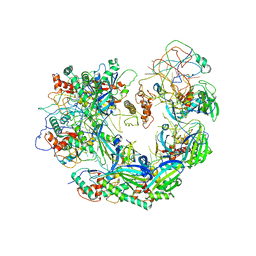 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | Descriptor: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQX
 
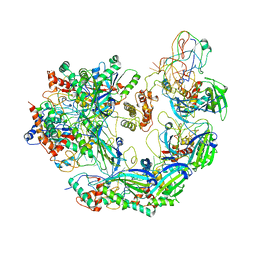 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQW
 
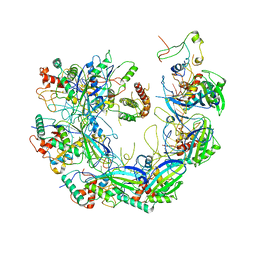 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1F71
 
 | |
1F70
 
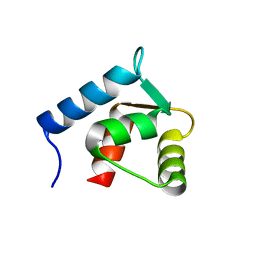 | |
1PZV
 
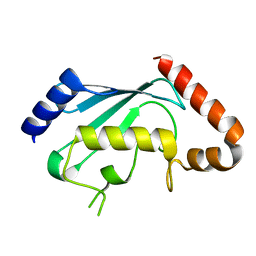 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1FNK
 
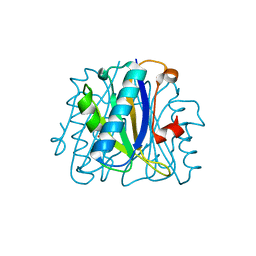 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88K/R90S | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1FNJ
 
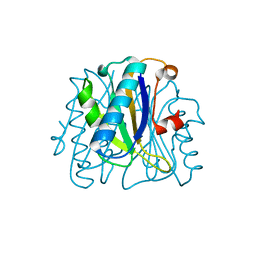 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88S/R90K | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1H53
 
 | | Binding of Phosphate and Pyrophosphate ions at the active site of human Angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, CITRIC ACID, PHOSPHATE ION | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.M, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
1H52
 
 | | Binding of Phosphate and Pyrophosphate ions at the active site of human Angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, PYROPHOSPHATE 2- | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.M, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
1HBY
 
 | | Binding of Phosphate and Pyrophosphate ions at the active site of human angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.S, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-04-21 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
2W81
 
 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
2W80
 
 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | Descriptor: | RNA (135-MER) | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-2 RNA SL5 domain. | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
 | | MERS 5' proximal stem-loop 5, conformation 3 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
