3FE8
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand soaked at pH 4.0 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
4GRW
 
 | | Structure of a complex of human IL-23 with 3 Nanobodies (Llama vHHs) | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, Nanobody 124C4, ... | | Authors: | Desmyter, A, Spinelli, S, Button, C, Saunders, M, de Haard, H, Rommelaere, H, Union, A, Cambillau, C. | | Deposit date: | 2012-08-27 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Neutralization of Human Interleukin 23 by Multivalent Nanobodies Explained by the Structure of Cytokine-Nanobody Complex.
Front Immunol, 8, 2017
|
|
3RX9
 
 | | 3D structure of SciN from an Escherichia coli Patotype | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Felisberto-Rodrigues, C, Durand, E, Aschtgen, M.-S, Blangy, S, Ortiz-Lombardia, M, Douzy, B, Cambillau, C, Cascales, E. | | Deposit date: | 2011-05-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Escherichia coli Pathovar.
Plos Pathog., 7, 2011
|
|
2X53
 
 | | Structure of the phage p2 baseplate in its activated conformation with Sr | | Descriptor: | ORF15, ORF16, PUTATIVE RECEPTOR BINDING PROTEIN, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6IC5
 
 | | Human cathepsin-C in complex with dipeptidyl cyclopropyl nitrile inhibitor 2 | | Descriptor: | (2~{S})-2-azanyl-~{N}-[(1~{R},2~{R})-1-(iminomethyl)-2-[4-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]phenyl]cyclopropyl]-3-thiophen-2-yl-propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
4HEP
 
 | | Complex of lactococcal phage TP901-1 with a llama vHH (vHH17) binder (nanobody) | | Descriptor: | BPP, SULFATE ION, vHH17 domain | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HEM
 
 | | Llama vHH-02 binder of ORF49 (RBP) from lactococcal phage TP901-1 | | Descriptor: | Anti-baseplate TP901-1 Llama vHH 02, BPP | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EQQ
 
 | | Structure of Ltp, a superinfection exclusion protein from the Streptococcus thermophilus temperate phage TP-J34 | | Descriptor: | PHOSPHATE ION, Putative host cell surface-exposed lipoprotein, SULFATE ION | | Authors: | Bebeacua, C, Lorenzo, C, Blangy, S, Spinelli, S, Heller, K, Cambillau, C. | | Deposit date: | 2012-04-19 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of a superinfection exclusion lipoprotein from phage TP-J34 and identification of the tape measure protein as its target.
Mol.Microbiol., 89, 2013
|
|
1NZJ
 
 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | Descriptor: | Hypothetical protein yadB, ZINC ION | | Authors: | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | Deposit date: | 2003-02-18 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
4ZS7
 
 | | Structural mimicry of receptor interaction by antagonistic IL-6 antibodies | | Descriptor: | Interleukin-6, Llama Fab fragment 68F2 heavy chain, Llama Fab fragment 68F2 light chain | | Authors: | Blanchetot, C, De Jonge, N, Desmyter, A, Ongenae, N, Hofman, E, Klarenbeek, A, Sadi, A, Hultberg, A, Kretz-Rommel, A, Spinelli, S, Loris, R, Cambillau, C, de Haard, H. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
J.Biol.Chem., 291, 2016
|
|
5FQH
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQE
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-N-ACETYLGALACTOSAMINIDASE, BROMIDE ION, ... | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FR0
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQG
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQF
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
1OJ7
 
 | | STRUCTURAL GENOMICS, UNKNOWN FUNCTION CRYSTAL STRUCTURE OF E. COLI K-12 YQHD | | Descriptor: | 5,6-DIHYDROXY-NADP, BORIC ACID, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Perrier, S, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-07-03 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E.Coli Alcohol Dehydrogenase Yqhd: Evidence of a Covalently Modified Nadp Coenzyme
J.Mol.Biol., 342, 2004
|
|
1ZRU
 
 | | structure of the lactophage p2 receptor binding protein in complex with glycerol | | Descriptor: | GLYCEROL, lactophage p2 receptor binding protein | | Authors: | Spinelli, S, Tremblay, D.M, Tegoni, M, Blangy, S, Huyghe, C, Desmyter, A, Labrie, S, de Haard, H, Moineau, S, Cambillau, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-22 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site.
J.Bacteriol., 188, 2006
|
|
5M30
 
 | | Structure of TssK from T6SS EAEC in complex with nanobody nb18 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Type VI secretion protein | | Authors: | Nguyen, V.S, Cambillau, C, Spinelli, C, Desmyter, A, Legrand, P, Cascales, E. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-21 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
6F27
 
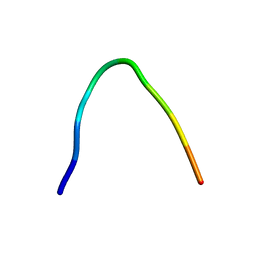 | | NMR solution structure of non-bound [des-Arg10]-kallidin (DAKD) | | Descriptor: | DAKD | | Authors: | Richter, C, Jonker, H.R.A, Schwalbe, H, Joedicke, L, Mao, J, Kuenze, G, Reinhart, C, Kalavacherla, T, Meiler, J, Preu, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
2XF6
 
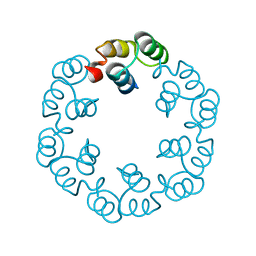 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XF7
 
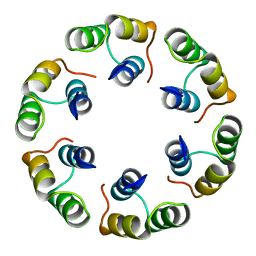 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. High-resolution structure. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XC8
 
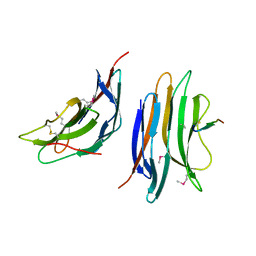 | | Crystal structure of the gene 22 product of the Bacillus subtilis SPP1 phage | | Descriptor: | GENE 22 PRODUCT | | Authors: | Veesler, D, Blangy, S, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp22 Shares Fold Similarity with a Domain of Lactococcal Phage P2 Rbp.
Protein Sci., 19, 2010
|
|
1W8G
 
 | | CRYSTAL STRUCTURE OF E. COLI K-12 YGGS | | Descriptor: | HYPOTHETICAL UPF0001 PROTEIN YGGS, ISOCITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sulzenbacher, G, Gruez, A, Spinelli, S, Roig-Zamboni, V, Pagot, F, Bignon, C, Vincentelli, R, Cambillau, C. | | Deposit date: | 2004-09-21 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E. Coli K-12 Yggs
To be Published
|
|
1O8C
 
 | | CRYSTAL STRUCTURE OF E. COLI K-12 YHDH WITH BOUND NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YHDH | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-26 | | Release date: | 2004-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia Coli Yhdh, a Putative Quinone Oxidoreductase
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1O89
 
 | | Crystal structure of E. COLI K-12 yhdH | | Descriptor: | YHDH | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-26 | | Release date: | 2004-02-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Escherichia Coli Yhdh, a Putative Quinone Oxidoreductase
Acta Crystallogr.,Sect.D, 60, 2004
|
|
