6YC0
 
 | | Crystal structure of the steady-state-SMX activated state of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YBZ
 
 | | Crystal structure of the D116N mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YC3
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6Y9Z
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,9) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YJ5
 
 | |
6Y9V
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-8,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9Y
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-7,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YBY
 
 | | Crystal structure of the D116N mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.6 | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YC1
 
 | | Crystal structure of the H30A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YC4
 
 | | Crystal structure of the steady-state activated state of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6Y9W
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,8) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7UG0
 
 | | TBOA-bound GltPh RSMR mutant in IFS state | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGX
 
 | | Asp-bound GltPh RSMR mutant in IFS-B1 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19 F NMR and Cryo-EM.
J.Am.Chem.Soc., 145, 2023
|
|
7UH6
 
 | | Asp-bound GltPh RSMR mutant in IFS-B2 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGJ
 
 | | TBOA-bound GltPh RSMR mutant in OFS state | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
8Q8H
 
 | |
7UGV
 
 | | Asp-bound GltPh RSMR mutant in IFS-A2 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19 F NMR and Cryo-EM.
J.Am.Chem.Soc., 145, 2023
|
|
7UH3
 
 | | Asp-bound GltPh RSMR mutant in iOFS state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGD
 
 | | Asp-bound GltPh RSMR mutant in IFS-A1 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
6ZDJ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YT4
 
 | | Crystal structure of the N112A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Maliar, N, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the N112A Mutant of the Light-Driven Sodium Pump KR2
Crystals, 2020
|
|
6YC2
 
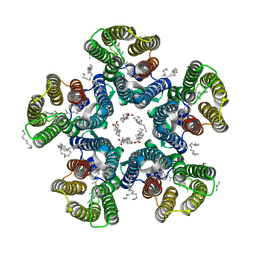 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ALANINE, EICOSANE, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
8QQZ
 
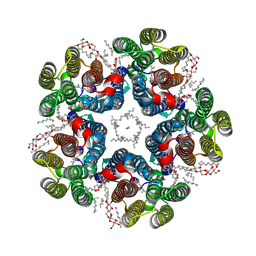 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QR0
 
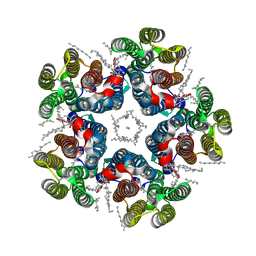 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QLE
 
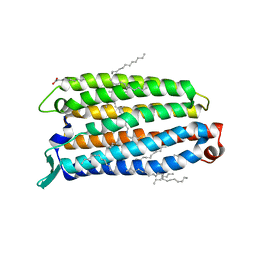 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 4.6 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
