1IFW
 
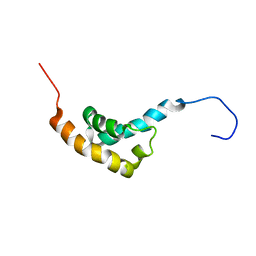 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN OF POLY(A) BINDING PROTEIN FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | POLYADENYLATE-BINDING PROTEIN, CYTOPLASMIC AND NUCLEAR | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Sprules, T, Ekiel, I, Gehring, K. | | Deposit date: | 2001-04-13 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein.
J.Biol.Chem., 277, 2002
|
|
1JDQ
 
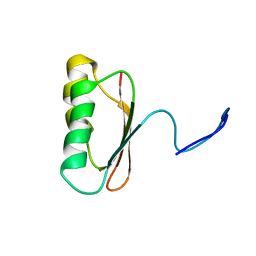 | | Solution Structure of TM006 Protein from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0983 | | Authors: | Denisov, A.Y, Finak, G, Yee, A, Kozlov, G, Gehring, K, Arrowsmith, C.H. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JCU
 
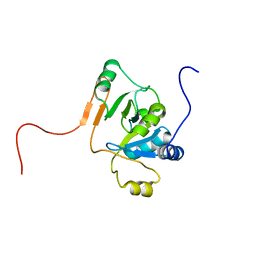 | |
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L1P
 
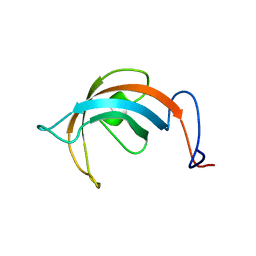 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | Descriptor: | trigger factor | | Authors: | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-02-19 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
1ME1
 
 | | Chimeric hairpin with 2',5'-linked RNA loop and RNA stem | | Descriptor: | 5'-R(*GP*GP*AP*CP*(U25)P*(U25)P*(C25)P*(5GP)P*GP*UP*CP*C)-3' | | Authors: | Denisov, A.Y, Hannoush, R.N, Gehring, K, Damha, M.J. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Motif Based on the Structure of Unusually Stable 2',5'-Linked r(UUCG) Loops
J.AM.CHEM.SOC., 125, 2003
|
|
1MK3
 
 | | SOLUTION STRUCTURE OF HUMAN BCL-W PROTEIN | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Denisov, A.Y, Madiraju, M.S, Chen, G, Khadir, A, Beauparlant, P, Attardo, G, Shore, G.C, Gehring, K. | | Deposit date: | 2002-08-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix
J.BIOL.CHEM., 278, 2003
|
|
1ME0
 
 | | Chimeric hairpin with 2',5'-linked RNA loop and DNA stem | | Descriptor: | DNA/RNA (5'-D(*GP*GP*AP*C)-R(P*(U25)P*(U25)P*(C25)P*(5GP))-D(P*GP*TP*CP*C)-3') | | Authors: | Denisov, A.Y, Hannoush, R.N, Gehring, K, Damha, M.J. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Motif Based on the Structure of Unusually Stable 2',5'-Linked r(UUCG) Loops
J.AM.CHEM.SOC., 125, 2003
|
|
1NEE
 
 | | Structure of archaeal translation factor aIF2beta from Methanobacterium thermoautrophicum | | Descriptor: | Probable translation initiation factor 2 beta subunit, ZINC ION | | Authors: | Gutierrez, P, Trempe, J.F, Siddiqui, N, Arrowsmith, C, Gehring, K. | | Deposit date: | 2002-12-11 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the archaeal translation initiation factor aIF2beta from Methanobacterium thermoautotrophicum: Implications for translation initiation.
Protein Sci., 13, 2004
|
|
1NMR
 
 | | Solution Structure of C-terminal Domain from Trypanosoma cruzi Poly(A)-Binding Protein | | Descriptor: | poly(A)-binding protein | | Authors: | Siddiqui, N, Kozlov, G, D'Orso, I, Trempe, J.F, Frasch, A.C.C, Gehring, K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain from poly(A)-binding protein in Trypanosoma cruzi: A vegetal PABC domain
Protein Sci., 12, 2003
|
|
7PVD
 
 | |
7PUY
 
 | |
4F02
 
 | |
2N7E
 
 | |
2IMT
 
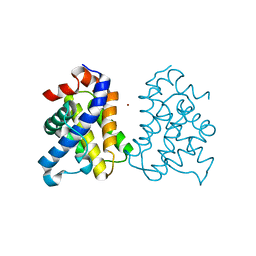 | | The X-ray Structure of a Bak Homodimer Reveals an Inhibitory Zinc Binding Site | | Descriptor: | Apoptosis regulator BAK, ZINC ION | | Authors: | Moldoveanu, T, Liu, Q, Tocilj, A, Watson, M, Shore, G.C, Gehring, K.B. | | Deposit date: | 2006-10-04 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site.
Mol.Cell, 24, 2006
|
|
2IMS
 
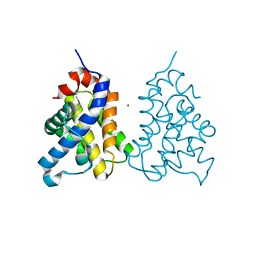 | | The X-ray Structure of a Bak Homodimer Reveals an Inhibitory Zinc Binding Site | | Descriptor: | Apoptosis regulator BAK, ZINC ION | | Authors: | Moldoveanu, T, Liu, Q, Tocilj, A, Watson, M, Shore, G.C, Gehring, K.B. | | Deposit date: | 2006-10-04 | | Release date: | 2006-12-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The X-Ray Structure of a BAK Homodimer Reveals an Inhibitory Zinc Binding Site
Mol.Cell, 24, 2006
|
|
2KNB
 
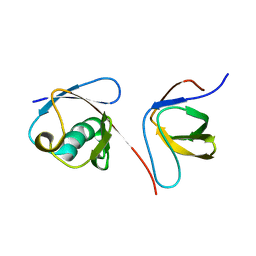 | | Solution NMR structure of the parkin Ubl domain in complex with the endophilin-A1 SH3 domain | | Descriptor: | E3 ubiquitin-protein ligase parkin, Endophilin-A1 | | Authors: | Trempe, J, Guennadi, K, Edna, C.M, Kalle, G. | | Deposit date: | 2009-08-20 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
2DYD
 
 | |
1EJE
 
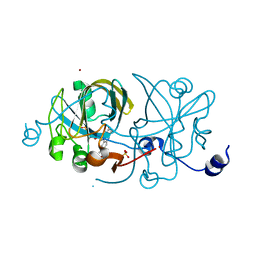 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
1EIJ
 
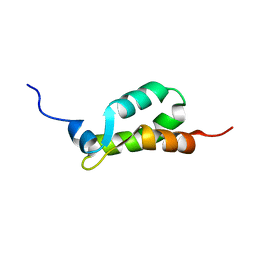 | | NMR ENSEMBLE OF METHANOBACTERIUM THERMOAUTOTROPHICUM PROTEIN 1615 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1615 | | Authors: | Christendat, D, Booth, V, Gernstein, M, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
2M5B
 
 | | The NMR structure of the BID-BAK complex | | Descriptor: | Bcl-2 homologous antagonist/killer, human_BID_BH3_SAHB | | Authors: | Moldoveanu, T, Grace, C.R, Kriwacki, R.W, Green, D.R. | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | BID-induced structural changes in BAK promote apoptosis.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1JRM
 
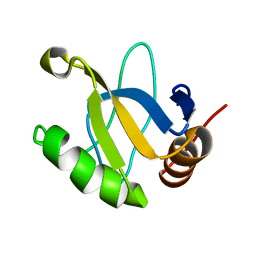 | |
1JW3
 
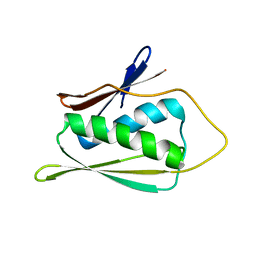 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW2
 
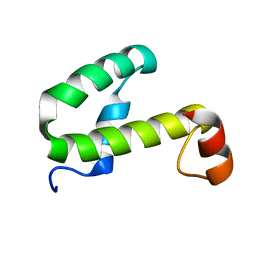 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2FH0
 
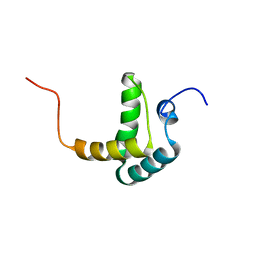 | |
