5YBI
 
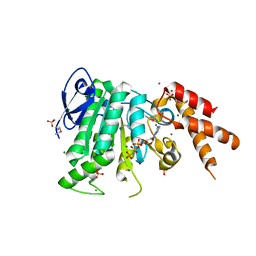 | | Structure of the bacterial pathogens ATPase with substrate AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Mu, Z.X, Gao, X.P, Cui, S. | | Deposit date: | 2017-09-05 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri.
Front Microbiol, 9, 2018
|
|
5ZT1
 
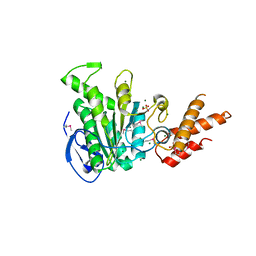 | | Structure of the bacterial pathogens ATPase with substrate ATP gamma S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Gao, X.P, Mu, Z.X, Cui, S. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.114 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri
Front Microbiol, 9, 2018
|
|
6JQX
 
 | |
6JQW
 
 | |
5FBO
 
 | | BTK-inhibitor co-structure | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-[8-azanyl-3-[(3~{R},6~{S})-1-cyclopropylcarbonyl-6-methyl-piperidin-3-yl]imidazo[1,5-a]pyrazin-1-yl]-3-fluoranyl-~{N}-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Discovery of 8-Amino-imidazo[1,5-a]pyrazines as Reversible BTK Inhibitors for the Treatment of Rheumatoid Arthritis.
ACS Med Chem Lett, 7, 2016
|
|
6YF1
 
 | | FKBP12 in complex with the BMP potentiator compound 8 at 1.12A resolution | | Descriptor: | (1aR,3R,5S,6R,7S,9R,10R,17aS,20S,21R,22S,25R,25aR)-25-Ethyl-10,22-dihydroxy-20-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-5,7-dimethoxy-1a,3,9,21-tetramethyloctadecahydro-2H-6,10-epoxyoxireno[p]pyrido[2,1-c][1,4]oxazacyclotricosine-11,12,18,24(1aH,14H)-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF2
 
 | | FKBP12 in complex with the BMP potentiator compound 6 at 1.03A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-23,25-dimethoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14-bis(oxidanyl)-17-(2-oxidanylidenepropyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF0
 
 | | FKBP12 in complex with the BMP potentiator compound 9 at 1.55 A resolution | | Descriptor: | 18-HYDROXYASCOMYCIN, Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF3
 
 | | FKBP12 in complex with the BMP potentiator compound 10 at 1.00A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-17-ethyl-25-methoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14,23-tris(oxidanyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
4WUM
 
 | |
5JEM
 
 | | Complex of IRF-3 with CBP | | Descriptor: | CREB-binding protein, Interferon regulatory factor 3 | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JER
 
 | | Structure of Rotavirus NSP1 bound to IRF-3 | | Descriptor: | Interferon regulatory factor 3, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEJ
 
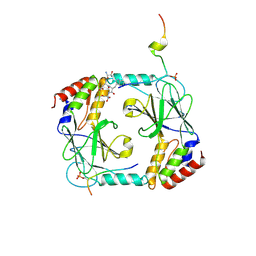 | | Phosphorylated STING in complex with IRF-3 CTD | | Descriptor: | Interferon regulatory factor 3, Stimulator of interferon genes protein | | Authors: | Li, P, Shu, C. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEL
 
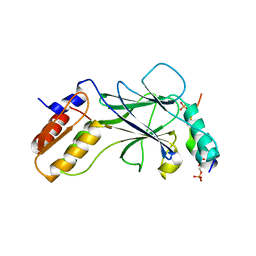 | | Phosphorylated TRIF in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, Phosphorylated TRIF peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6FKQ
 
 | | THE CRYSTAL STRUCTURE OF A FRAGMENT OF NETRIN-1 IN COMPLEX WITH A FRAGMENT OF DRAXIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bhowmick, T, Meijers, R. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron, 97, 2018
|
|
4DN5
 
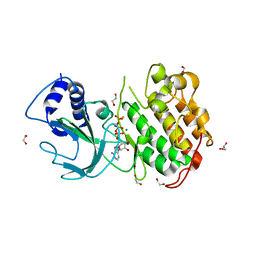 | | Crystal Structure of NF-kB-inducing Kinase (NIK) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Min, X, Liu, J, Sudom, A, Walker, N.P, Wang, Z. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Nuclear Factor Kappa B-inducing kinase domain reveals a constitutively active conformation
J.Biol.Chem., 287, 2012
|
|
5XGQ
 
 | |
4S00
 
 | |
4RZY
 
 | |
4S01
 
 | |
3NFC
 
 | |
149D
 
 | |
7KPT
 
 | | Crystal structure of CtdE in complex with FAD and substrate 4 | | Descriptor: | (6aR,7aS,11S,13aS)-6,6,11-trimethyl-4-(3-methylbut-2-en-1-yl)-6,6a,7,8,9,10,11,14-octahydro-5H,13H-13a,7a-(epiminomethano)quinolizino[2,3-b]carbazol-16-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
7KPQ
 
 | | Crystal structure of CtdE in complex with FAD | | Descriptor: | FAD-dependent monooxygenase CtdE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
6X3P
 
 | | Co-structure of BTK kinase domain with L-005298385 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{8-amino-3-[(6R,8aS)-3-oxooctahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-(cyclopropyloxy)-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
