3TW4
 
 | | Crystal Structure of Human Septin 7 GTPase Domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Septin-7 | | Authors: | Serrao, V.H.B, Alessandro, F, Pereira, H.M, Thiemann, O.T, Garratt, R.C. | | Deposit date: | 2011-09-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Promiscuous interactions of human septins: The GTP binding domain of SEPT7 forms filaments within the crystal.
Febs Lett., 585, 2011
|
|
3UMF
 
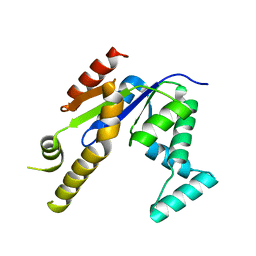 | |
3UL5
 
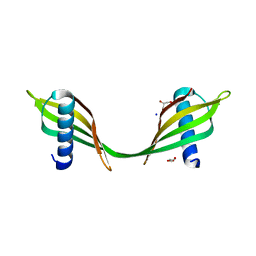 | | Saccharum officinarum canecystatin-1 in space group C2221 | | Descriptor: | Canecystatin-1, GLYCEROL, SODIUM ION | | Authors: | Valadares, N.F, Pereira, H.M, Oliveira-Silva, R, Garratt, R.C. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallography and NMR studies of domain-swapped canecystatin-1.
Febs J., 280, 2013
|
|
7KEH
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
3SS4
 
 | |
3SS3
 
 | |
3SS5
 
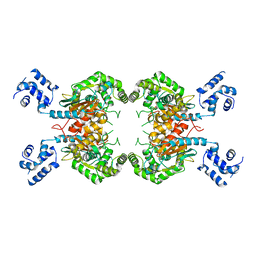 | |
5JX4
 
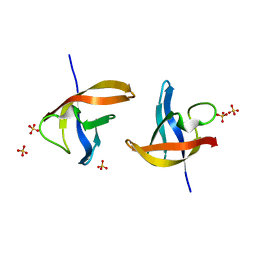 | | Crystal structure of E36-G37del mutant of the Bacillus caldolyticus cold shock protein. | | Descriptor: | Cold shock protein CspB, SULFATE ION | | Authors: | Carvajal, A, Castro-Fernandez, V, Cabrejos, D, Fuentealba, M, Pereira, H.M, Vallejos, G, Cabrera, R, Garratt, R.C, Komives, E.A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual dimerization of a BcCsp mutant leads to reduced conformational dynamics.
FEBS J., 284, 2017
|
|
5F28
 
 | | Crystal structure of FAT domain of Focal Adhesion Kinase (FAK) bound to the transcription factor MEF2C | | Descriptor: | Focal adhesion kinase 1, MEF2C | | Authors: | Cardoso, A.C, Ambrosio, A.L.B, Dessen, A, Franchini, K.G. | | Deposit date: | 2015-12-01 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | FAK Forms a Complex with MEF2 to Couple Biomechanical Signaling to Transcription in Cardiomyocytes.
Structure, 24, 2016
|
|
