3VUK
 
 | | Crystal structure of a cysteine-deficient mutant M5 in MAP kinase JNK1 | | Descriptor: | Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1, SULFATE ION | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-07-02 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
3VUI
 
 | | Crystal structure of a cysteine-deficient mutant M2 in MAP kinase JNK1 | | Descriptor: | Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1, SULFATE ION | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-06-28 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
3VUG
 
 | | Crystal structure of a cysteine-deficient mutant M2 in MAP kinase JNK1 | | Descriptor: | Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1, SULFATE ION | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-06-28 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
3WE7
 
 | | Crystal Structure of Diacetylchitobiose Deacetylase from Pyrococcus horikoshii | | Descriptor: | ACETIC ACID, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Mine, S, Nakamura, T, Fukuda, Y, Inoue, T, Uegaki, K, Sato, T. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-07 | | Last modified: | 2014-08-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
3WII
 
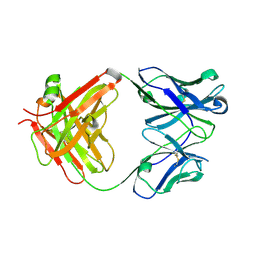 | | Crystal structure of the Fab fragment of B2212A, a murine monoclonal antibody specific for the third fibronectin domain (Fn3) of human ROBO1. | | Descriptor: | anti-human ROBO1 antibody B2212A Fab heavy chain, anti-human ROBO1 antibody B2212A Fab light chain | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, S, Nakakido, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3WIH
 
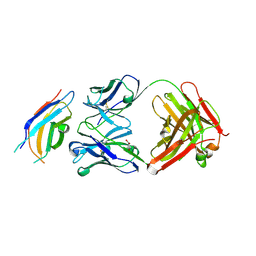 | | Crystal structure of the third fibronectin domain (Fn3) of human ROBO1 in complex with the Fab fragment of murine monoclonal antibody B2212A. | | Descriptor: | GLYCEROL, Roundabout homolog 1, anti-human ROBO1 antibody B2212A Fab heavy chain, ... | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3WYQ
 
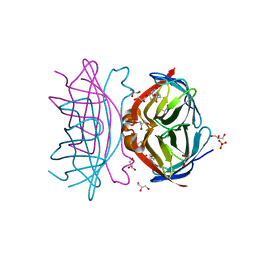 | | Crystal structure of the low-immunogenic core streptavidin mutant LISA-314 (Y22S/Y83S/R84K/E101D/R103K/E116N) at 1.0 A resolution | | Descriptor: | BIOTIN, GLYCEROL, SULFATE ION, ... | | Authors: | Kawato, T, Mizohata, E, Meshizuka, T, Doi, H, Kawamura, T, Matsumura, H, Yumura, K, Tsumoto, K, Kodama, T, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of streptavidin mutant with low immunogenicity.
J.Biosci.Bioeng., 119, 2015
|
|
3WYP
 
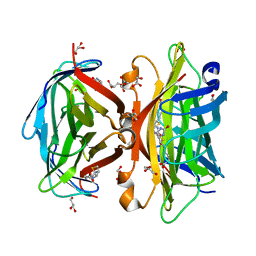 | | Crystal structure of wild-type core streptavidin in complex with D-biotin/biotin-D-sulfoxide at 1.3 A resolution | | Descriptor: | BIOTIN, BIOTIN-D-SULFOXIDE, GLYCEROL, ... | | Authors: | Kawato, T, Mizohata, E, Meshizuka, T, Doi, H, Kawamura, T, Matsumura, H, Yumura, K, Tsumoto, K, Kodama, T, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of streptavidin mutant with low immunogenicity.
J.Biosci.Bioeng., 119, 2015
|
|
3WZN
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin at 1.3 A resolution | | Descriptor: | BIOTIN, SULFATE ION, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3A2V
 
 | | Peroxiredoxin (C207S) from Aeropyrum pernix K1 complexed with hydrogen peroxide | | Descriptor: | PEROXIDE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Ishikawa, K, Matsumura, H, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3A5W
 
 | | Peroxiredoxin (wild type) from Aeropyrum pernix K1 (reduced form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, T, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-08-12 | | Release date: | 2009-10-27 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3VI2
 
 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | Descriptor: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | Authors: | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
2Z5G
 
 | | Crystal structure of T1 lipase F16L mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, Thermostable lipase, ... | | Authors: | Matsumura, H, Yamamoto, T, Inoue, T, Kai, Y. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
3AXM
 
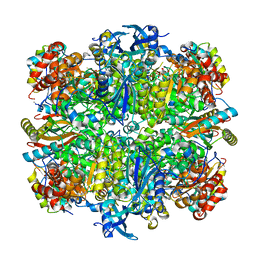 | | Structure of rice Rubisco in complex with 6PG | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
3AXK
 
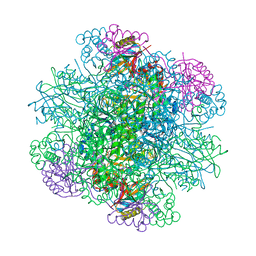 | | Structure of rice Rubisco in complex with NADP(H) | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
3A2X
 
 | | Peroxiredoxin (C50S) from Aeropyrum pernix K1 (acetate-bound form) | | Descriptor: | ACETATE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3A2W
 
 | | Peroxiredoxin (C50S) from Aeropytum pernix K1 (peroxide-bound form) | | Descriptor: | GLYCEROL, PEROXIDE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3B20
 
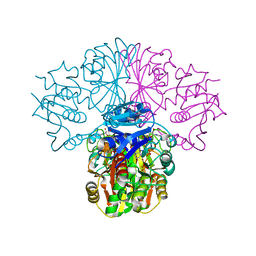 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with NADfrom Synechococcus elongatus" | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Matsumura, H, Kai, A, Maeda, T, Inoue, T. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
3B1K
 
 | |
3B1J
 
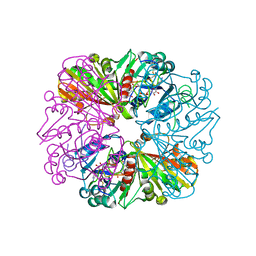 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with CP12 in the presence of copper from Synechococcus elongatus | | Descriptor: | COPPER (II) ION, CP12, Glyceraldehyde 3-phosphate dehydrogenase (NADP+), ... | | Authors: | Matsumura, H, Kai, A, Inoue, T. | | Deposit date: | 2011-07-04 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
3AGU
 
 | | Hemerythrin-like domain of DcrH (semimet-R) | | Descriptor: | CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Onoda, A, Okamoto, Y, Sugimoto, H, Mizohata, E, Inoue, T, Shiro, Y, Hayashi, T. | | Deposit date: | 2010-04-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Characteristics of Diiron Site with Large Cavity in Hemerythrin-like Domain of DcrH
to be published
|
|
3AGT
 
 | | Hemerythrin-like domain of DcrH (met) | | Descriptor: | CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Onoda, A, Okamoto, Y, Sugimoto, H, Mizohata, E, Inoue, T, Shiro, Y, Hayashi, T. | | Deposit date: | 2010-04-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characteristics of Diiron Site with Large Cavity in Hemerythrin-like Domain of DcrH
to be published
|
|
3AGV
 
 | | Crystal structure of a human IgG-aptamer complex | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*(UFT)P*GP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*GP*AP*AP*A*GP*GP*AP*AP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*A)-3', CALCIUM ION, Ig gamma-1 chain C region, ... | | Authors: | Nomura, Y, Sugiyama, S, Sakamoto, T, Miyakawa, S, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Nakamura, Y, Matsumura, H. | | Deposit date: | 2010-04-08 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational plasticity of RNA for target recognition as revealed by the 2.15 A crystal structure of a human IgG-aptamer complex
Nucleic Acids Res., 38, 2010
|
|
3VP9
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
2ZA3
 
 | |
