7CBM
 
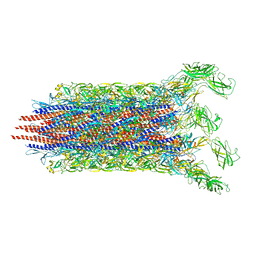 | | Cryo-EM structure of the flagellar distal rod with partial hook from Salmonella | | Descriptor: | Flagellar basal-body rod protein FlgF, Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG0
 
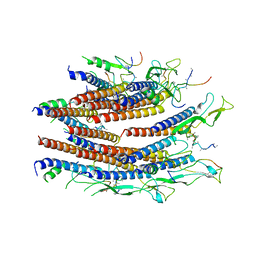 | | Cryo-EM structure of the flagellar proximal rod with FliF peptides from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
8JMV
 
 | |
8JMW
 
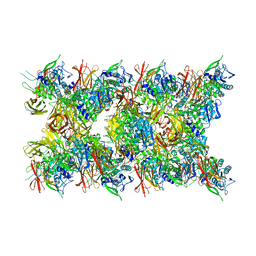 | | Fibril form of serine peptidase Vpr | | Descriptor: | S8 family serine peptidase | | Authors: | Cao, Q, Cheng, Y. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Serine peptidase Vpr forms enzymatically active fibrils outside Bacillus bacteria revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
8T1Y
 
 | |
8T1Z
 
 | |
8SIR
 
 | |
8SIS
 
 | |
8SDG
 
 | |
8SDF
 
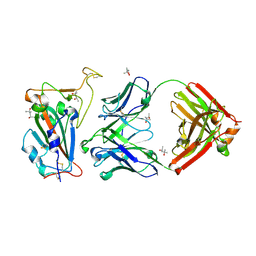 | |
8SIT
 
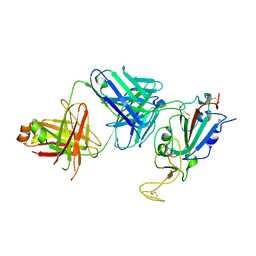 | |
8SIQ
 
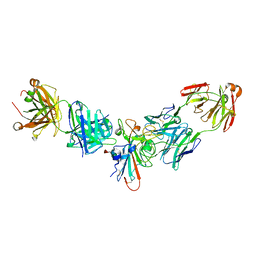 | |
8SDH
 
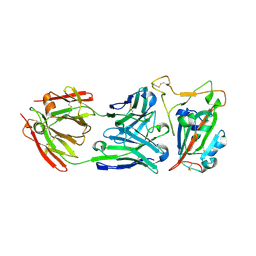 | |
5YGI
 
 | |
5YVK
 
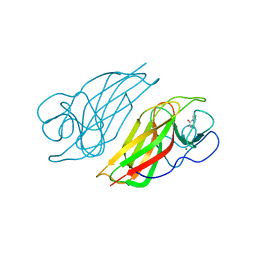 | | Crystal structure of a cyclase Famc1 from Fischerella ambigua UTEX 1903 | | Descriptor: | CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium, cyclase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.292 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YVP
 
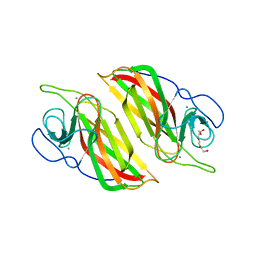 | | Crystal structure of an apo form cyclase Filc1 from Fischerella sp. TAU | | Descriptor: | CALCIUM ION, TETRAETHYLENE GLYCOL, cyclase A | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z54
 
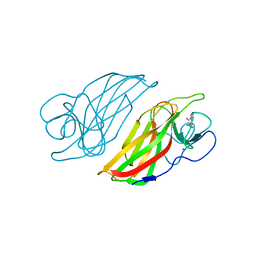 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-17 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z53
 
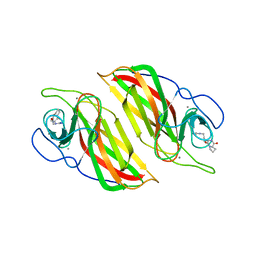 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole U synthase, CALCIUM ION, SULFATE ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YVL
 
 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, cyclase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5ZFJ
 
 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with 4-(1H-Indol-3-yl)butan-2-one | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(1~{H}-indol-3-yl)butan-2-one, CALCIUM ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5TPT
 
 | |
6CBV
 
 | | Crystal structure of BRIL bound to an affinity matured synthetic antibody. | | Descriptor: | BRIL, FORMIC ACID, GLYCEROL, ... | | Authors: | Mukherjee, S, Skrobek, B, Kossiakoff, A.A. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | Descriptor: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | Descriptor: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8V
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
