2FHF
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltotetraose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
5N7W
 
 | | Computationally designed functional antibody | | Descriptor: | Antibody Fragment Heavy Chain, Antibody Fragment Light Chain, Interleukin-17A | | Authors: | Hargreaves, D, Breed, J. | | Deposit date: | 2017-02-21 | | Release date: | 2018-11-14 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Computational Design of Epitope-Specific Functional Antibodies.
Cell Rep, 25, 2018
|
|
6SHC
 
 | | Crystal structure of human IRE1 luminal domain Q105C | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Unstructured regions in IRE1 alpha specify BiP-mediated destabilisation of the luminal domain dimer and repression of the UPR.
Elife, 8, 2019
|
|
5N8G
 
 | | Serial Cu nitrite reductase structures at elevated cryogenic temperature, 240K. Dataset 2. | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Horrell, S, Kekilli, D, Hough, M, Strange, R. | | Deposit date: | 2017-02-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Active-site protein dynamics and solvent accessibility in native Achromobacter cycloclastes copper nitrite reductase.
IUCrJ, 4, 2017
|
|
6SDV
 
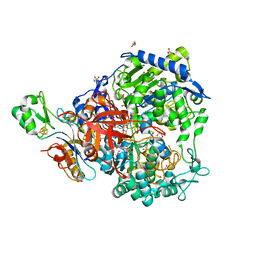 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Formate reduced form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
1NW1
 
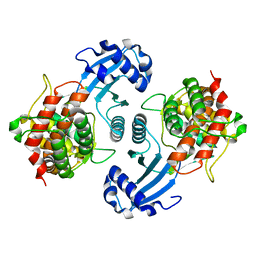 | | Crystal Structure of Choline Kinase | | Descriptor: | CALCIUM ION, Choline kinase (49.2 kD) | | Authors: | Peisach, D, Gee, P, Kent, C, Xu, Z. | | Deposit date: | 2003-02-05 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of Choline Kinase Reveals a Eukaryotic Protein Kinase Fold
Structure, 11, 2003
|
|
3VT1
 
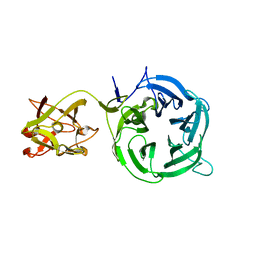 | | Crystal structure of Ct1,3Gal43A in complex with galactose | | Descriptor: | Ricin B lectin, beta-D-galactopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
5NA1
 
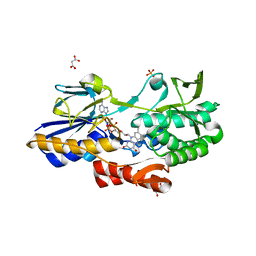 | | NADH:quinone oxidoreductase (NDH-II) from Staphylococcus aureus - holoprotein structure - 2.32 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, NADH dehydrogenase-like protein SAOUHSC_00878, ... | | Authors: | Brito, J.A, Athayde, D, Sousa, F.M, Sena, F.V, Pereira, M.M, Archer, M. | | Deposit date: | 2017-02-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The key role of glutamate 172 in the mechanism of type II NADH:quinone oxidoreductase of Staphylococcus aureus.
Biochim. Biophys. Acta, 1858, 2017
|
|
5NB3
 
 | | High resolution C-phycoerythrin from marine cyanobacterium Phormidium sp. A09DM at pH 7.5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Sonani, R.R, Roszak, A.W, Ortmann de Percin Northumberland, C, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An improved crystal structure of C-phycoerythrin from the marine cyanobacterium Phormidium sp. A09DM.
Photosyn. Res., 135, 2018
|
|
2FM0
 
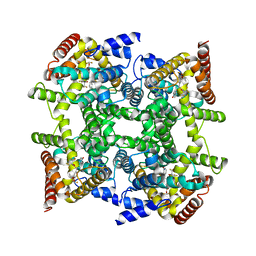 | | Crystal structure of PDE4D in complex with L-869298 | | Descriptor: | (S)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHY L)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
2FH1
 
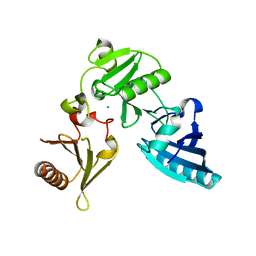 | | C-terminal half of gelsolin soaked in low calcium at pH 4.5 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
5NK2
 
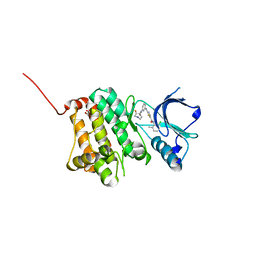 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2b | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
1NOW
 
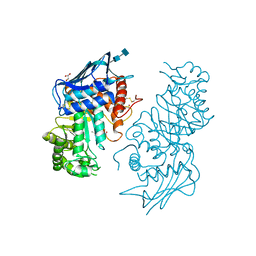 | | Human lysosomal beta-hexosaminidase isoform B in complex with (2R,3R,4S,5R)-2-Acetamido-3,4-Dihydroxy-5-Hydroxymethyl-Piperidinium Chloride (GalNAc-isofagomine) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
5NKA
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2g | | Descriptor: | 1,2-ETHANEDIOL, 4-[[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylamino]cyclohexane-1-carboxylic acid, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6S9F
 
 | | Drosophila OTK, extracellular domains 3-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tyrosine-protein kinase-like otk | | Authors: | Rozbesky, D, Jones, E.Y. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Drosophila OTK Is a Glycosaminoglycan-Binding Protein with High Conformational Flexibility.
Structure, 28, 2020
|
|
6S9X
 
 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor 15c | | Descriptor: | RAC-alpha serine/threonine-protein kinase, ~{N}-[3-[1-[[4-[5-[(4-hydroxyphenyl)methyl]-6-oxidanylidene-2-phenyl-1~{H}-pyrazin-3-yl]phenyl]methyl]piperidin-4-yl]-2-oxidanylidene-1~{H}-benzimidazol-5-yl]propanamide | | Authors: | Landel, I, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent-Allosteric Inhibitors to Achieve Akt Isoform-Selectivity.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
2EUH
 
 | | HOLO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE COMPLEX WITH NADP+ | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
2FE0
 
 | |
5NJ6
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | Descriptor: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NJZ
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1g | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-(piperidin-4-ylcarbamoyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NIK
 
 | | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump | | Descriptor: | Macrolide export ATP-binding/permease protein MacB, Macrolide export protein MacA, Outer membrane protein TolC | | Authors: | Fitzpatrick, A.W.P, Llabres, S, Neuberger, A, Blaza, J.N, Bai, X.-C, Okada, U, Murakami, S, van Veen, H.W, Zachariae, U, Scheres, S.H.W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump.
Nat Microbiol, 2, 2017
|
|
5NK1
 
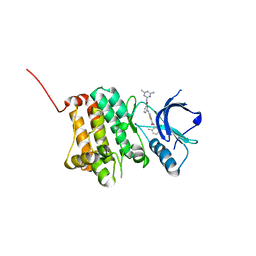 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1k | | Descriptor: | 2-[[3-[(4-azanyl-6-methyl-1,3,5-triazin-2-yl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6SG9
 
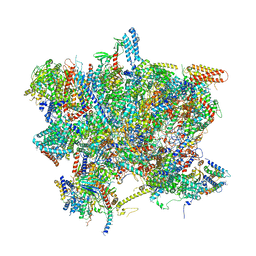 | | Head domain of the mt-SSU assemblosome from Trypanosoma brucei | | Descriptor: | 9S rRNA, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Saurer, M, Ramrath, D.J.F, Niemann, M, Calderaro, S, Prange, C, Mattei, S, Scaiola, A, Leitner, A, Bieri, P, Horn, E.K, Leibundgut, M, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery.
Science, 365, 2019
|
|
5NK6
 
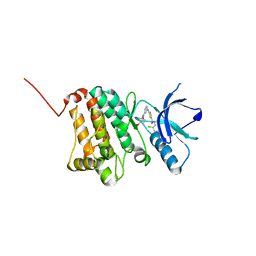 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2d | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[(4-oxidanylcyclohexyl)carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.267 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NK5
 
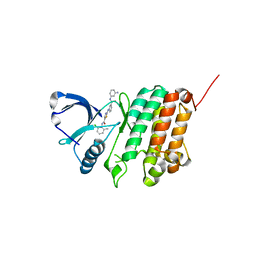 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1m | | Descriptor: | 2-[[3-[(3-aminophenyl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
