2W9D
 
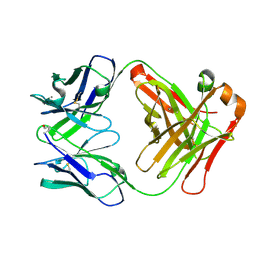 | | Structure of Fab fragment of the ICSM 18 - anti-Prp therapeutic antibody at 1.57 A resolution. | | Descriptor: | CALCIUM ION, ICSM 18-ANTI-PRP THERAPEUTIC FAB HEAVY CHAIN, ICSM 18-ANTI-PRP THERAPEUTIC FAB LIGHT CHAIN | | Authors: | Antonyuk, S.V, Trevitt, C.R, Strange, R.W, Jackson, G.S, Sangar, D, Batchelor, M, Jones, S, Georgiou, T, Cooper, S, Fraser, C, Khalili-Shirazi, A, Clarke, A.R, Hasnain, S.S, Collinge, J. | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of Human Prion Protein Bound to a Therapeutic Antibody.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5OGF
 
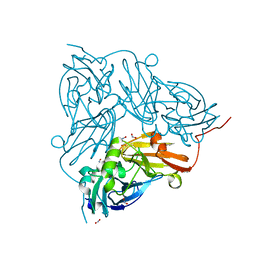 | | Cu nitrite reductase serial data at varying temperatures RT 0.27MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
2WZ0
 
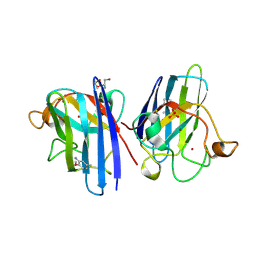 | | L38V SOD1 mutant complexed with aniline. | | Descriptor: | 2-METHOXY-5-METHYLANILINE, COPPER (II) ION, D-SERINE, ... | | Authors: | Antonyuk, S, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-20 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
2WZ6
 
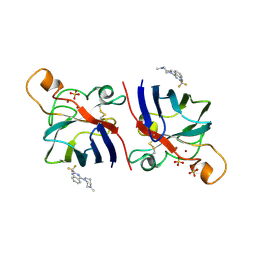 | | G93A SOD1 mutant complexed with Quinazoline. | | Descriptor: | 4-(4-METHYL-1,4-DIAZEPAN-1-YL)-2-(TRIFLUOROMETHYL)QUINAZOLINE, COPPER (II) ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
2WZ5
 
 | | L38V SOD1 mutant complexed with L-methionine. | | Descriptor: | COPPER (II) ION, METHIONINE, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
2WYZ
 
 | | L38V SOD1 mutant complexed with UMP | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-20 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
2WQK
 
 | | Crystal Structure of Sure Protein from Aquifex aeolicus | | Descriptor: | 5'-NUCLEOTIDASE SURE, SODIUM ION, SULFATE ION | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-08-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Sure Protein from Aquifex Aeolicus Vf5 at 1.5 A Resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5OG5
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.21MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5OF6
 
 | | Cu nitrite reductase serial data at varying temperatures 190K 0.48MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5OFG
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.06MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5OG2
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.09MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
2VR8
 
 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.36 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2YV3
 
 | | Crystal Structure of Aspartate Semialdehyde Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Kagawa, W, Fujikawa, N, Kurumizaka, H, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-07 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Aspartate Semialdehyde Dehydrogenase from Thermus thermophilus HB8
To be Published
|
|
1E30
 
 | |
7JOR
 
 | | Neutron structure of ferric Dehaloperoxidase B | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Carey, L.M, Ghiladi, R.A, Meilleur, F, Myles, D.A.A. | | Deposit date: | 2020-08-07 | | Release date: | 2021-09-08 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.05 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
4Y9M
 
 | | PA3825-EAL Metal-Free-Apo Structure | | Descriptor: | PA3825-EAL, PHOSPHATE ION | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
4Y9P
 
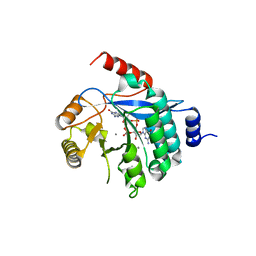 | | PA3825-EAL Ca-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, PA3825-EAL | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | MucR and PA3825 EAL-phosphodiesterase domains from Pseudomonas aeruginosa suggest roles for three metals in the active site
To Be Published
|
|
7KCU
 
 | | Joint neutron/X-ray structure of Oxyferrous Dehaloperoxidase B | | Descriptor: | Dehaloperoxidase B, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Carey, L.M, Ghiladi, R.A, Meilleur, F, Myles, D. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
7KFM
 
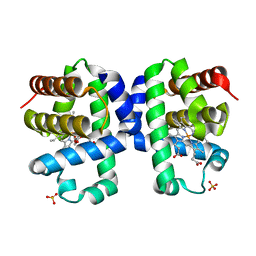 | | Room temperature oxyferrous Dehaloperoxidase B | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Dehaloperoxidase B, OXYGEN MOLECULE, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
5MF5
 
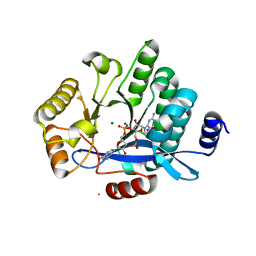 | | PA3825-EAL Mg-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Diguanylate phosphodiesterase, MAGNESIUM ION | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
5MFU
 
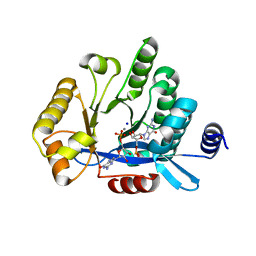 | | PA3825-EAL Mn-pGpG Structure | | Descriptor: | Diguanylate phosphodiesterase, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-11-18 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
5MKG
 
 | | PA3825-EAL Ca-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Diguanylate phosphodiesterase | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
7ADQ
 
 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno-Chicano, T.M, Ebrahim, A, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | Deposit date: | 2020-09-15 | | Release date: | 2021-11-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
5YTU
 
 | |
5YTO
 
 | |
