2R5D
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HIV entry inhibitor PIE7, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RF3
 
 | | Crystal Structure of Tricyclo-DNA: An Unusual Compensatory Change of Two Adjacent Backbone Torsion Angles | | Descriptor: | 5'-d(CGCG(TCY)ATTCGCG)-3', SPERMINE, ZINC ION | | Authors: | Pallan, P.S, Ittig, D, Heroux, A, Wawrzak, Z, Leumann, C.J, Egli, M. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of tricyclo-DNA: an unusual compensatory change of two adjacent backbone torsion angles.
Chem.Commun.(Camb.), 2008, 2008
|
|
1WYY
 
 | | Post-fusion hairpin conformation of the sars coronavirus spike glycoprotein | | Descriptor: | CHLORIDE ION, E2 Glycoprotein | | Authors: | Duquerroy, S, Vigouroux, A, Rottier, P.J.M, Rey, F.A, Bosch, B.J. | | Deposit date: | 2005-02-18 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein
Virology, 335, 2005
|
|
3LA6
 
 | | Octameric kinase domain of the E. coli tyrosine kinase Wzc with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Tyrosine-protein kinase wzc | | Authors: | Gruszczyk, J, Nessler, S, Gueguen-Chaignon, V, Vigouroux, A, Bechet, E, Grangeasse, C. | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export
Mol.Microbiol., 77, 2010
|
|
1R7S
 
 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5E8B
 
 | |
5EMX
 
 | |
2VF1
 
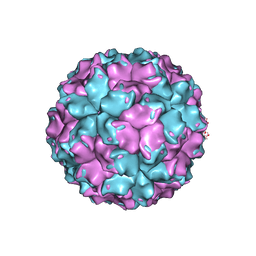 | | X-ray crystallographic structure of the picobirnavirus capsid | | Descriptor: | CAPSID PROTEIN | | Authors: | Duquerroy, S, Da Costa, B, Vigouroux, A, Lepault, J, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Picobirnavirus Crystal Structure Provides Functional Insights Into Virion Assembly and Cell Entry.
Embo J., 28, 2009
|
|
5E3F
 
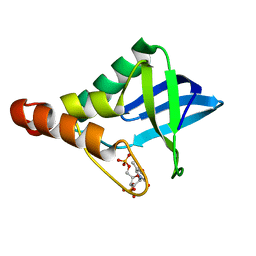 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92K at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Caro, J.A, Heroux, A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-10-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92K at cryogenic temperature
To be Published
|
|
2R0V
 
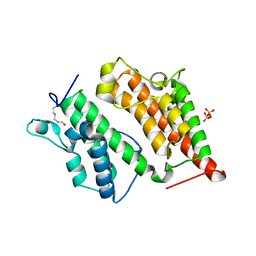 | | Structure of the Rsc4 tandem bromodomain acetylated at K25 | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, SULFATE ION | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
5F4Y
 
 | |
2R3C
 
 | | Structure of the gp41 N-peptide in complex with the HIV entry inhibitor PIE1 | | Descriptor: | CHLORIDE ION, HIV entry inhibitor PIE1, YTTRIUM (III) ION, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-08-29 | | Release date: | 2007-10-02 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5B
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | HIV entry inhibitor PIE7, SULFATE ION, gp41 N-peptide | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5F5P
 
 | | Molecular Basis for Shroom2 Recognition by Rock1 | | Descriptor: | CHLORIDE ION, Protein Shroom2, Rho-associated protein kinase 1 | | Authors: | Zalewski, J.K, VanDemark, A.P, Heroux, A. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.568 Å) | | Cite: | Structure of the Shroom-Rho Kinase Complex Reveals a Binding Interface with Monomeric Shroom That Regulates Cell Morphology and Stimulates Kinase Activity.
J. Biol. Chem., 291, 2016
|
|
1YWM
 
 | | Crystal structure of the N-terminal domain of group B Streptococcus alpha C protein | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, C protein alpha-antigen, GLYCEROL | | Authors: | Auperin, T.C, Bolduc, G.R, Baron, M.J, Heroux, A, Filman, D.J, Madoff, L.C, Hogle, J.M. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the N-terminal domain of the group B streptococcus alpha C protein.
J.Biol.Chem., 280, 2005
|
|
4XGK
 
 | | Crystal structure of UDP-galactopyranose mutase from Corynebacterium diphtheriae in complex with 2-[4-(4-chlorophenyl)-7-(2-thienyl)-2-thia-5,6,8,9-tetrazabicyclo[4.3.0]nona-4,7,9-trien-3-yl]acetic | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | Authors: | Wangkanont, K, Heroux, A, Forest, K.T, Kiessling, L.L. | | Deposit date: | 2014-12-31 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Virtual Screening for UDP-Galactopyranose Mutase Ligands Identifies a New Class of Antimycobacterial Agents.
Acs Chem.Biol., 10, 2015
|
|
6QAK
 
 | |
4Y3I
 
 | | PAS-GAF fragment from Deinococcus radiodurans BphP assembled with BV - Y307S, low dose | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, F, Burgie, E.S, Yu, T, Heroux, A, Schatz, G.C, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-20 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
J.Am.Chem.Soc., 137, 2015
|
|
6QAP
 
 | |
6HLY
 
 | | Structure in P212121 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, agropinic acid | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HLZ
 
 | | Structure in C2 form of the PBP AgtB from A.tumefacien R10 in complex with agropinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Agropine permease, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HM2
 
 | | Structure in P1 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, SODIUM ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
4YGL
 
 | | NaClO4--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | Carbonic anhydrase 2, HYDROXIDE ION, PERCHLORATE ION, ... | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
6HLX
 
 | | Structure of the PBP AgaA in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6QAO
 
 | |
