6JDO
 
 | |
6JDA
 
 | |
6JDB
 
 | |
6JDH
 
 | |
1O7M
 
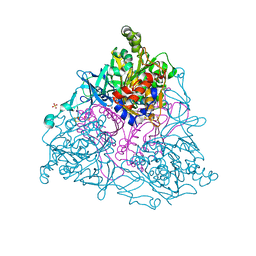 | | NAPHTHALENE 1,2-DIOXYGENASE, BINARY COMPLEX WITH DIOXYGEN | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-11 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1QFJ
 
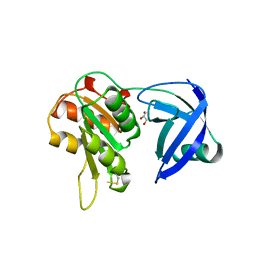 | | CRYSTAL STRUCTURE OF NAD(P)H:FLAVIN OXIDOREDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, PROTEIN (FLAVIN REDUCTASE) | | Authors: | Ingelman, M, Ramaswamy, S, Niviere, V, Fontecave, M, Eklund, H. | | Deposit date: | 1999-04-12 | | Release date: | 1999-06-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli.
Biochemistry, 38, 1999
|
|
1O7P
 
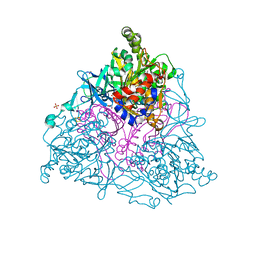 | | NAPHTHALENE 1,2-DIOXYGENASE, PRODUCT COMPLEX | | Descriptor: | (1R, 2S)-CIS 1,2 DIHYDROXY-1,2-DIHYDRONAPHTHALENE, 1,2-ETHANEDIOL, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-11 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1O7N
 
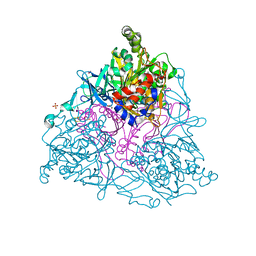 | | NAPHTHALENE 1,2-DIOXYGENASE, TERNARY COMPLEX WITH DIOXYGEN AND INDOLE | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-11 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1O7G
 
 | | NAPHTHALENE 1,2-DIOXYGENASE WITH NAPHTHALENE BOUND IN THE ACTIVE SITE. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-05 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1O7H
 
 | | NAPHTHALENE 1,2-DIOXYGENASE WITH OXIDIZED RIESKE IRON SULPHUR CENTER SITE. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-05 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1O7W
 
 | | NAPHTHALENE 1,2-DIOXYGENASE, FULLY REDUCED FORM | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-14 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1R0R
 
 | | 1.1 Angstrom Resolution Structure of the Complex Between the Protein Inhibitor, OMTKY3, and the Serine Protease, Subtilisin Carlsberg | | Descriptor: | CALCIUM ION, Ovomucoid, subtilisin carlsberg | | Authors: | Horn, J.R, Ramaswamy, S, Murphy, K.P. | | Deposit date: | 2003-09-22 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and energetics of protein-protein interactions: the role of conformational heterogeneity in OMTKY3 binding to serine proteases
J.Mol.Biol., 331, 2003
|
|
1R26
 
 | | Crystal structure of thioredoxin from Trypanosoma brucei brucei | | Descriptor: | Thioredoxin | | Authors: | Friemann, R, Schmidt, H, Ramaswamy, S, Forstner, M, Krauth-Siegel, R.L, Eklund, H. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of thioredoxin from Trypanosoma brucei brucei
FEBS Lett., 554, 2003
|
|
1CDO
 
 | |
6R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441D MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
7O3K
 
 | |
8B01
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc | | Descriptor: | DECANE, DOCOSANE, HEXANE, ... | | Authors: | Davies, J.S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2022-09-06 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
8F0Y
 
 | | Lipocalin-like Milk protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein | | Authors: | Subramanian, R, KanagaVijayan, D, Shantakumar, R.P.S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
7Q02
 
 | | Zn-free structure of lipocalin-like Milk protein, inspired from Diploptera punctata, expressed in Saccharomyces cerevisiae | | Descriptor: | Milk protein, PALMITOLEIC ACID | | Authors: | Banerjee, S, Dhanabalan, K.V, Ramaswamy, S. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of recombinantly expressed cockroach Lili-Mip protein in glycosylated and deglycosylated forms.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
3KZD
 
 | |
3KZE
 
 | |
6B16
 
 | | P21-activated kinase 1 in complex with a 4-azaindole inhibitor | | Descriptor: | N~4~-(5-cyclopropyl-1H-pyrazol-3-yl)-N~2~-[(1S)-1-(1H-pyrrolo[3,2-b]pyridin-5-yl)ethyl]pyrimidine-2,4-diamine, SULFATE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Rouge, L, Wang, W. | | Deposit date: | 2017-09-16 | | Release date: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Synthesis and evaluation of a series of 4-azaindole-containing p21-activated kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
6NYH
 
 | | Structure of human RIPK1 kinase domain in complex with GNE684 | | Descriptor: | (5S)-N-[(3S)-7-methoxy-1-methyl-2-oxo-2,3,4,5-tetrahydro-1H-pyrido[3,4-b]azepin-3-yl]-5-phenyl-6,7-dihydro-5H-pyrrolo[1,2-b][1,2,4]triazole-2-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RIP1 inhibition blocks inflammatory diseases but not tumor growth or metastases.
Cell Death Differ., 27, 2020
|
|
7QHA
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol | | Descriptor: | Megabody c7HopQ, Putative TRAP-type C4-dicarboxylate transport system, large permease component, ... | | Authors: | North, R.A, Davies, J.S, Morado, D, Dobson, R.C.J. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
5HVY
 
 | | CDK8/CYCC IN COMPLEX WITH COMPOUND 20 | | Descriptor: | CHLORIDE ION, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Kiefer, J.R, Schneider, E.V, Maskos, K, Bergeron, P, Koehler, M. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Development of a Series of Potent and Selective Type II Inhibitors of CDK8.
Acs Med.Chem.Lett., 7, 2016
|
|
