7CJV
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a dilute environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
3A1A
 
 | | Crystal Structure of the DNMT3A ADD domain | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, ZINC ION | | Authors: | Otani, J, Arita, K, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2009-03-28 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Embo Rep., 10, 2009
|
|
7CJW
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a crowded environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
2ZKD
 
 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), ... | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
3ATQ
 
 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, TETRADECANE | | Authors: | Sasaki, D, Fujihashi, M, Murakami, M, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
7D8K
 
 | | Solution structure of the methyl-CpG binding domain of MBD6 from Arabidopsis thaliana | | Descriptor: | Methyl-CpG-binding domain-containing protein 6 | | Authors: | Mahana, Y, Ohki, I, Walinda, E, Morimoto, D, Sugase, K, Shirakawa, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Methylated DNA Recognition by the Methyl-CpG Binding Domain of MBD6 from Arabidopsis thaliana .
Acs Omega, 7, 2022
|
|
3B0F
 
 | | Crystal structure of the UBA domain of p62 and its interaction with ubiquitin | | Descriptor: | SULFATE ION, Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2ZKE
 
 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DAP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DAP*DG)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
3A4R
 
 | | The crystal structure of SUMO-like domain 2 in Nip45 | | Descriptor: | 1,2-ETHANEDIOL, NFATC2-interacting protein, SULFATE ION | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
1IXF
 
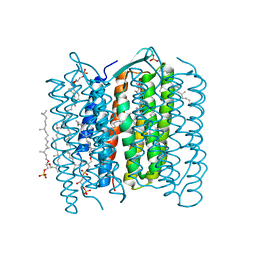 | | Crystal Structure of the K intermediate of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Matsui, Y, Sakai, K, Murakami, M, Shiro, Y, Adachi, S, Okumura, H, Kouyama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-20 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary Photoreaction of Bacteriorhodopsin
J.MOL.BIOL., 324, 2002
|
|
1EHD
 
 | |
1IQB
 
 | |
2RRI
 
 | | NMR structure of vasoactive intestinal peptide in DPC Micelle | | Descriptor: | Vasoactive intestinal peptide | | Authors: | Umetsu, Y, Tenno, T, Goda, N, Shirakawa, M, Ikegami, T, Hiroaki, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural difference of vasoactive intestinal peptide in two distinct membrane-mimicking environments
Biochim.Biophys.Acta, 1814, 2011
|
|
2RQQ
 
 | | Structure of C-terminal region of Cdt1 | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Jee, J.G, Mizuno, T, Kamada, K, Tochio, H, Hiroaki, H, Hanaoka, F, Shirakawa, M. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins
J.Biol.Chem., 285, 2010
|
|
2RPA
 
 | | The solution structure of N-terminal domain of microtubule severing enzyme | | Descriptor: | Katanin p60 ATPase-containing subunit A1 | | Authors: | Iwaya, N, Kuwahara, Y, Unzai, S, Nagata, T, Tomii, K, Goda, N, Tochio, H, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2008-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A common substrate recognition mode conserved between katanin P60 and VPS4 governs microtubule severing and membrane skeleton reorganization
J.Biol.Chem., 285, 2010
|
|
1JSE
 
 | |
2MX2
 
 | | UBX-L domain of VCIP135 | | Descriptor: | Deubiquitinating protein VCIP135 | | Authors: | Iwazu, T, Murayama, S, Igarashi, R, Hrioaki, H, Shirakawa, M, Tochio, H. | | Deposit date: | 2014-12-07 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction mode of the UBX-L domain of VCIP135 determined by solution NMR spectroscopy
To be Published
|
|
1FR0
 
 | | SOLUTION STRUCTURE OF THE HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ANAEROBIC SENSOR KINASE ARCB FROM ESCHERICHIA COLI. | | Descriptor: | ARCB | | Authors: | Ikegami, T, Okada, T, Ohki, I, Hirayama, J, Mizuno, T, Shirakawa, M. | | Deposit date: | 2000-09-07 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamic character of the histidine-containing phosphotransfer domain of anaerobic sensor kinase ArcB from Escherichia coli.
Biochemistry, 40, 2001
|
|
1COP
 
 | |
1COO
 
 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
1JSF
 
 | |
1EHH
 
 | |
1UEL
 
 | | Solution structure of ubiquitin-like domain of hHR23B complexed with ubiquitin-interacting motif of proteasome subunit S5a | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, UV excision repair protein RAD23 homolog B | | Authors: | Fujiwara, K, Tenno, T, Jee, J.G, Sugasawa, K, Ohki, I, Kojima, C, Tochio, H, Hiroaki, H, Hanaoka, H, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ubiquitin-interacting Motif of S5a Bound to the Ubiquitin-like Domain of HR23B
J.Biol.Chem., 279, 2004
|
|
1WLF
 
 | | Structure of the N-terminal domain of PEX1 AAA-ATPase: Characterization of a putative adaptor-binding domain | | Descriptor: | Peroxisome biogenesis factor 1, SULFATE ION | | Authors: | Shiozawa, K, Maita, N, Tomii, K, Seto, A, Goda, N, Tochio, H, Akiyama, Y, Shimizu, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the N-terminal Domain of PEX1 AAA-ATPase: CHARACTERIZATION OF A PUTATIVE ADAPTOR-BINDING DOMAIN
J.Biol.Chem., 279, 2004
|
|
1X0S
 
 | | Crystal structure of the 13-cis isomer of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nishikawa, T, Murakami, M, Kouyama, T. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the 13-cis isomer of bacteriorhodopsin in the dark-adapted state.
J.Mol.Biol., 352, 2005
|
|
