6B9X
 
 | | Crystal structure of Ragulator | | Descriptor: | Hepatitis B virus x interacting protein, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | SU, M.-Y, Hurley, J.H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex.
Mol. Cell, 68, 2017
|
|
8GNI
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GNJ
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GQ5
 
 | |
8PQ2
 
 | | XBB 1.0 RBD bound to P4J15 (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, P4J15 Fragment Antigen-Binding Heavy Chain, P4J15 Fragment Antigen-Binding Light Chain, ... | | Authors: | Duhoo, Y, Lau, K. | | Deposit date: | 2023-07-10 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
8PSD
 
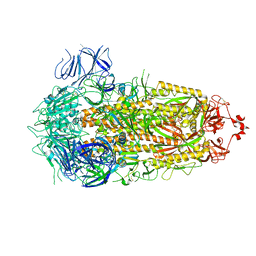 | | SARS-CoV-2 XBB 1.0 closed conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Duhoo, Y, Lau, K. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
8SEM
 
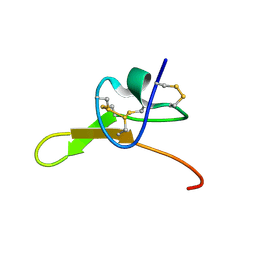 | |
8SED
 
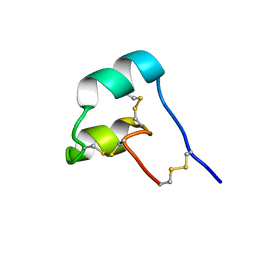 | |
6BUC
 
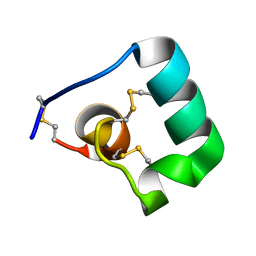 | |
2K9O
 
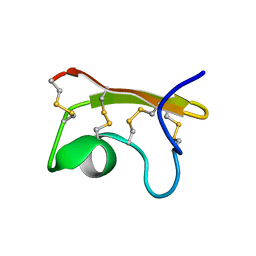 | | Solution structure of Vm24 synthetic scorpion toxin | | Descriptor: | Vm24 SCORPION toxin | | Authors: | del Rio-Portilla, F, Hernandez-Lopez, R, Possani-Postay, L, Gurrola, G. | | Deposit date: | 2008-10-20 | | Release date: | 2009-11-03 | | Last modified: | 2014-05-28 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and chemical synthesis of Vaejovis mexicanus peptide 24: a novel potent blocker of Kv1.3 potassium channels of human T lymphocytes.
Biochemistry, 51, 2012
|
|
3IQX
 
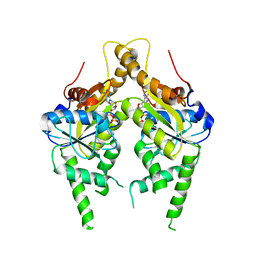 | | ADP complex of C.therm. Get3 in closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IQW
 
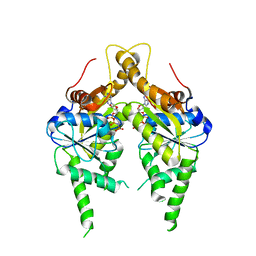 | | AMPPNP complex of C. therm. Get3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3P9K
 
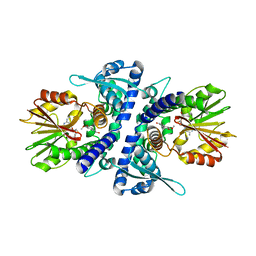 | |
3P9C
 
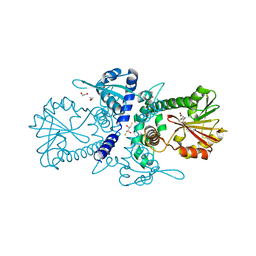 | | Crystal structure of perennial ryegrass LpOMT1 bound to SAH | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
6UX5
 
 | |
8TT9
 
 | | X-ray structure of Macrophage Migration Inhibitory Factor (MIF) Covalently Bound to 4-hydroxyphenylpyruvate (HPP) | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor | | Authors: | Schroder, G.C, Meilleur, F, Nix, J.C, Crichlow, G.V, Lolis, E.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray structure of Macrophage Migration Inhibitory Factor (MIF) Covalently Bound to 4-hydroxyphenylpyruvate (HPP)
To Be Published
|
|
3P9I
 
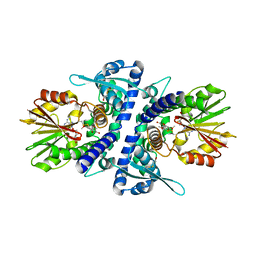 | | Crystal structure of perennial ryegrass LpOMT1 complexed with S-adenosyl-L-homocysteine and sinapaldehyde | | Descriptor: | (2E)-3-(4-hydroxy-3,5-dimethoxyphenyl)prop-2-enal, BETA-MERCAPTOETHANOL, Caffeic acid O-methyltransferase, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
6VGJ
 
 | | N-terminal variant of CXCL13 | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Rosenberg Jr, E.M, Lolis, E.J. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The N-terminal length and side-chain composition of CXCL13 affect crystallization, structure and functional activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8W4J
 
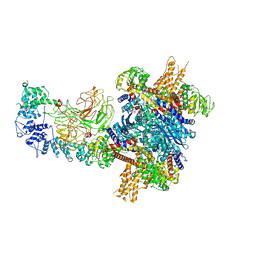 | |
8GRW
 
 | | Spiroplasma melliferum FtsZ F224M bound to GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.40005422 Å) | | Cite: | Dynamics of interdomain rotation facilitates FtsZ filament assembly.
J.Biol.Chem., 300, 2024
|
|
6CYT
 
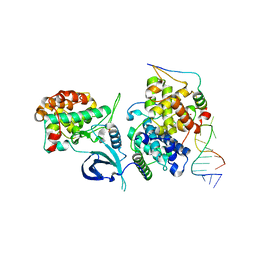 | | HIV-1 TAR loop in complex with Tat:AFF4:P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze Gahmen, U, Hurley, J.H. | | Deposit date: | 2018-04-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
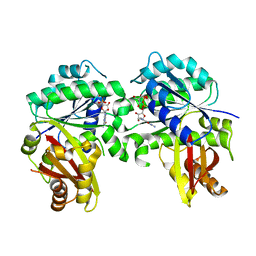
6MC1
 
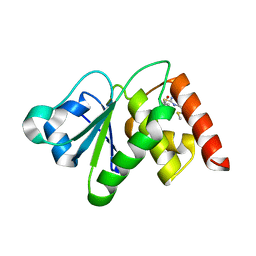 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
6MZT
 
 | | Solution structure of alpha-KTx-6.21 (UroTx) from Urodacus yaschenkoi | | Descriptor: | Potassium channel toxin alpha-KTx 6.21 | | Authors: | Chin, Y.K.-Y, Luna-Ramirez, K, Anangi, R, King, G.F. | | Deposit date: | 2018-11-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the potency and selectivity of Urotoxin, a potent Kv1 blocker from scorpion venom.
Biochem. Pharmacol., 174, 2020
|
|
7U4F
 
 | |
7U4G
 
 | | Neuraminidase from influenza virus A/Shandong/9/1993(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Lei, R, Hernandez Garcia, A. | | Deposit date: | 2022-02-28 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prevalence and mechanisms of evolutionary contingency in human influenza H3N2 neuraminidase.
Nat Commun, 13, 2022
|
|
