4U81
 
 | | MEK1 Kinase bound to small molecule inhibitor G659 | | Descriptor: | 8-[(4-cyclopropyl-2-fluorophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-7-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Robarge, K.D, Ultsch, M.H, Weismann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U7Z
 
 | | Mitogen-Activated Protein Kinase Kinase (MEK1) bound to G805 | | Descriptor: | 5-[(4-bromo-2-chlorophenyl)amino]-4-fluoro-N-(2-hydroxyethoxy)-1-methyl-1H-benzimidazole-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Robarge, K.D, Ultsch, M.H, Wiesmann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U80
 
 | | MEK 1 kinase bound to G799 | | Descriptor: | 3-[(4-cyclopropyl-2-fluorophenyl)amino]-N-(2-hydroxyethoxy)furo[3,2-c]pyridine-2-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ultsch, M.H, Robarge, K.D, Weismann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5IME
 
 | | Crystal structure of P21-activated kinase 1 (PAK1) in complex with compound 9 | | Descriptor: | 8-(3-aminopropyl)-6-[2-chloro-4-(3-methyl-2-oxopyrazin-1(2H)-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | Authors: | Li, D, Wang, W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Chemically Diverse Group I p21-Activated Kinase (PAK) Inhibitors Impart Acute Cardiovascular Toxicity with a Narrow Therapeutic Window.
J.Med.Chem., 59, 2016
|
|
1V3A
 
 | |
2WCU
 
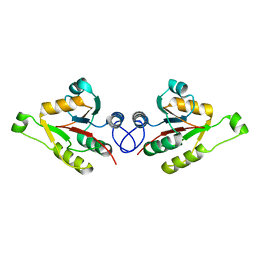 | | Crystal structure of mammalian FucU | | Descriptor: | PROTEIN FUCU HOMOLOG, alpha-L-fucopyranose | | Authors: | Lee, K.-H, Kim, M.-S, Suh, H.-Y, Ku, B, Song, Y.-L, Oh, B.-H. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures and Enzyme Mechanism of a Dual Fucose Mutarotase/Ribose Pyranase
J.Mol.Biol., 391, 2009
|
|
2P3T
 
 | |
2QHB
 
 | |
2CKX
 
 | |
1JDQ
 
 | | Solution Structure of TM006 Protein from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0983 | | Authors: | Denisov, A.Y, Finak, G, Yee, A, Kozlov, G, Gehring, K, Arrowsmith, C.H. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JCU
 
 | |
1JRM
 
 | |
1JW2
 
 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW3
 
 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2PLN
 
 | |
1OGF
 
 | | The Structure of Bacillus subtilis RbsD complexed with glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1OGE
 
 | | The Structure of Bacillus subtilis RbsD complexed with Ribose 5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, CHLORIDE ION, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1OGC
 
 | | The Structure of Bacillus subtilis RbsD complexed with D-ribose | | Descriptor: | CHLORIDE ION, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1OGD
 
 | | The Structure of Bacillus subtilis RbsD complexed with D-ribose | | Descriptor: | CHLORIDE ION, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD, beta-D-ribopyranose | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1YLA
 
 | | Ubiquitin-conjugating enzyme E2-25 kDa (Huntington interacting protein 2) | | Descriptor: | Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Choe, J, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Bochkarev, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
1AYM
 
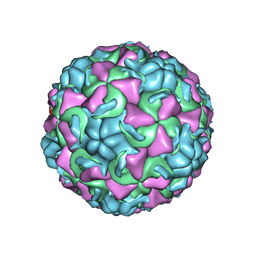 | | HUMAN RHINOVIRUS 16 COAT PROTEIN AT HIGH RESOLUTION | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle.
Structure, 5, 1997
|
|
6UYC
 
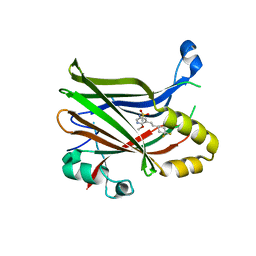 | | Crystal structure of TEAD2 bound to Compound 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-{5-[(E)-2-(4,4-difluorocyclohexyl)ethenyl]-6-methoxypyridin-3-yl}methanesulfonamide, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
1QJG
 
 | |
6UYB
 
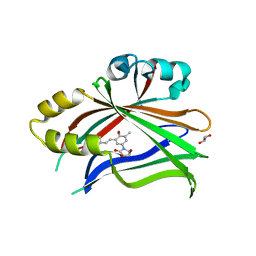 | | Crystal structure of TEAD2 bound to Compound 1 | | Descriptor: | (3R,4R)-1-{3-[(E)-2-(4-chlorophenyl)ethenyl]-4-methoxy-5-methylphenyl}-3,4-dihydroxypyrrolidin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
