6S6Q
 
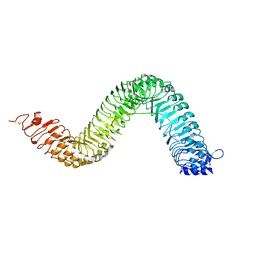 | | Crystal structure of the LRR ectodomain of the plant membrane receptor kinase GASSHO1/SCHENGEN3 from Arabidopsis thaliana in complex with CASPARIAN STRIP INTEGRITY FACTOR 2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LRR receptor-like serine/threonine-protein kinase GSO1, ... | | Authors: | Okuda, S, Moretti, A, Hothorn, M. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5A61
 
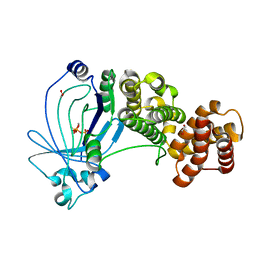 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two manganese ions. | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A5Y
 
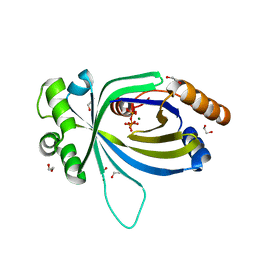 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A67
 
 | |
4LSX
 
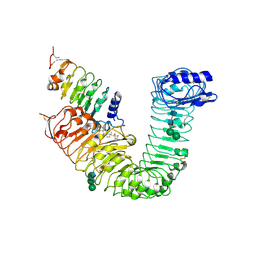 | | Plant steroid receptor ectodomain bound to brassinolide and SERK1 co-receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, ... | | Authors: | Santiago, J, Henzler, C, Hothorn, M. | | Deposit date: | 2013-07-23 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases.
Science, 341, 2013
|
|
4LSC
 
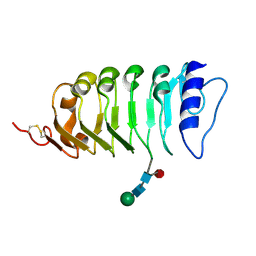 | | Isolated SERK1 co-receptor ectodomain at high resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Somatic embryogenesis receptor kinase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Santiago, J, Henzler, C, Hothorn, M. | | Deposit date: | 2013-07-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases.
Science, 341, 2013
|
|
2XNR
 
 | |
2XNQ
 
 | |
3BXJ
 
 | | Crystal Structure of the C2-GAP Fragment of synGAP | | Descriptor: | Ras GTPase-activating protein SynGAP | | Authors: | Pena, V, Hothorn, M, Eberth, A, Kaschau, N, Parret, A, Gremer, L, Bonneau, F, Ahmadian, M.R, Scheffzek, K. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction.
Embo Rep., 9, 2008
|
|
3GYL
 
 | | Structure of Prostasin at 1.3 Angstroms resolution in complex with a Calcium Ion. | | Descriptor: | CALCIUM ION, GLYCEROL, Prostasin | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations.
Protein Sci., 18, 2009
|
|
3FVF
 
 | | The Crystal Structure of Prostasin Complexed with Camostat at 1.6 Angstroms Resolution | | Descriptor: | 1-[4-(hydroxymethyl)phenyl]guanidine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2009-01-15 | | Release date: | 2009-05-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations.
Protein Sci., 18, 2009
|
|
3GYM
 
 | | Structure of Prostasin in Complex with Aprotinin | | Descriptor: | Pancreatic trypsin inhibitor, Prostasin | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations.
Protein Sci., 18, 2009
|
|
2OP3
 
 | | The structure of cathepsin S with a novel 2-arylphenoxyacetaldehyde inhibitor derived by the Substrate Activity Screening (SAS) method | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-[(2',3',4'-TRIFLUOROBIPHENYL-2-YL)OXY]ETHANOL, Cathepsin S, ... | | Authors: | Spraggon, G, Inagaki, H, Tsuruoka, H, Hornsby, M, Lesley, S.A, Ellman, J.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and optimization of selective, nonpeptidic inhibitors of cathepsin S with an unprecedented binding mode.
J.Med.Chem., 50, 2007
|
|
2HHN
 
 | | Cathepsin S in complex with non covalent arylaminoethyl amide. | | Descriptor: | Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2006-06-28 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HH5
 
 | | Crystal Structure of Cathepsin S in complex with a Zinc mediated non-covalent arylaminoethyl amide | | Descriptor: | CHLORIDE ION, Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, ... | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S. | | Deposit date: | 2006-06-27 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HXZ
 
 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor (Hexagonal spacegroup) | | Descriptor: | Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE, SULFATE ION | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
2H7J
 
 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor. | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
2FXK
 
 | | Crystal structure of the macro-domain of human core histone variant macroH2A1.1 (form A) | | Descriptor: | H2A histone family, member Y isoform 1 | | Authors: | Kustatscher, G, Hothorn, M, Pugieux, C, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2006-02-06 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Splicing regulates NAD metabolite binding to histone macroH2A.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8CC2
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, gallinamide A, ... | | Authors: | Rubesova, P, Brynda, J, Mares, M, Gerwick, W.H. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
8CCU
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1 | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, [(2~{S})-1-[[(2~{S})-1-[[(2~{S})-5-[(2~{S})-3-methoxy-2-(2-methylpropyl)-5-oxidanylidene-2~{H}-pyrrol-1-yl]-5-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl] (2~{S},3~{S})-2-(dimethylamino)-3-methyl-pentanoate | | Authors: | Rubesova, P, Brynda, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
8CD9
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 6 | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin B-like peptidase (C01 family), SODIUM ION, ... | | Authors: | Rubesova, P, Brynda, J, Fanfrlik, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
7NUU
 
 | | Crystal structure of human AMDHD2 in complex with Zn | | Descriptor: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Ruegenberg, S, Kroef, V, Baumann, U, Denzel, M.S. | | Deposit date: | 2021-03-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | GFPT2/GFAT2 and AMDHD2 act in tandem to control the hexosamine pathway.
Elife, 11, 2022
|
|
7NUT
 
 | | Crystal structure of human AMDHD2 in complex with Zn and GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Ruegenberg, S, Kroef, V, Baumann, U, Denzel, M.S. | | Deposit date: | 2021-03-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | GFPT2/GFAT2 and AMDHD2 act in tandem to control the hexosamine pathway.
Elife, 11, 2022
|
|
7QBN
 
 | | Structure of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBL
 
 | |
