9DRB
 
 | | Binary complex of DNA polymerase iota with product DNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*CP*T)-3'), DNA polymerase iota | | Authors: | Frevert, Z, Reusch, D, Freudenthal, B, Washington, M.T. | | Deposit date: | 2024-09-25 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Visualizing DNA polymerase iota catalyze Hoogsteen-directed DNA synthesis
Nat Commun, 16, 2025
|
|
9DRC
 
 | | Ternary substrate complex of DNA polymerase iota R71A mutant with DNA (template A) and dTTP | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA polymerase iota, ... | | Authors: | Frevert, Z, Reusch, D, Freudenthal, B, Washington, M.T. | | Deposit date: | 2024-09-25 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Visualizing DNA polymerase iota catalyze Hoogsteen-directed DNA synthesis
Nat Commun, 16, 2025
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6SH7
 
 | |
6RTE
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
6YLV
 
 | | Translation initiation factor 4E in complex with 4-Cl-Bn7GpppG mRNA 5' cap analog | | Descriptor: | 4-Cl-Bn7GpppG mRNA 5' cap analog, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Kubacka, D, Wojcik, R, Baranowski, M.R, Kowalska, J, Jemielity, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66005659 Å) | | Cite: | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
6TDB
 
 | | Neuropilin2-b1 domain in a complex with the C-terminal VEGFB167 peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, C-terminal VEGFB167 peptide, ... | | Authors: | Eldrid, C, Yu, L, Yelland, T, Fotinou, C, Djordjevic, S. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NRP2-b1 domain in a complex with the C-terminal VEGFB167 peptide
To Be Published
|
|
6T6X
 
 | | Structure of the Bottromycin epimerase BotH in complex with substrate | | Descriptor: | (4~{R})-2-[(1~{R})-1-[[(2~{S})-2-[[(2~{S})-3-methyl-2-[[(4~{Z},6~{S},9~{S},12~{S})-2,8,11-tris(oxidanylidene)-6,9-di(propan-2-yl)-1,4,7,10-tetrazabicyclo[10.3.0]pentadec-4-en-5-yl]amino]butanoyl]amino]-3-phenyl-propanoyl]amino]-3-oxidanyl-3-oxidanylidene-propyl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, BotH | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-20 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
8G6C
 
 | | GTP Cyclohydrolase-IB with manganese | | Descriptor: | 1,2-ETHANEDIOL, GTP cyclohydrolase FolE2, MANGANESE (II) ION | | Authors: | McWhorter, K.L, Amaya Lopez, C.Y, Davis, K.M. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Combatting melioidosis with chemical synthetic lethality.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7PUX
 
 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
7QIV
 
 | | Structure of human C3b in complex with the EWE nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Pedersen, H, Andersen, G.R. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Engineering of a Complement Component C3-Binding Nanobody Improves Specificity and Adds Cofactor Activity.
Front Immunol, 13, 2022
|
|
8V1J
 
 | |
8V1X
 
 | | Crystal structure of polyketide synthase (PKS) thioreductase domain from Streptomyces coelicolor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pereira, J.H, Dan, Q, Keasling, J, Adams, P.D. | | Deposit date: | 2023-11-21 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A polyketide-based biosynthetic platform for diols, amino alcohols and hydroxy acids
Nat Catal, 8, 2025
|
|
7Q7F
 
 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at atmospheric pressure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q7N
 
 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at 120 MPa helium gas pressure in a sapphire capillary | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q7G
 
 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at 30 MPa helium gas pressure in a sapphire capillary | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q7O
 
 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at atmospheric pressure after high helium gas pressure release | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q7J
 
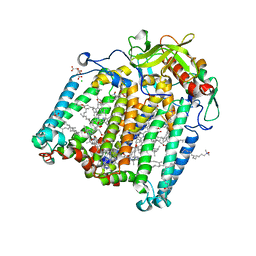 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at 75 MPa helium gas pressure in a sapphire capillary | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
8VJZ
 
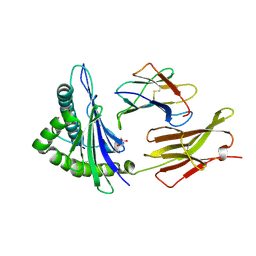 | | HLA-A*03:01 with WT KRAS-10mer | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GTPase KRas, ... | | Authors: | Sim, M.J.W, Sun, P.D. | | Deposit date: | 2024-01-08 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and structural characterization of a mutant KRAS-G12V specific TCR restricted by HLA-A3.
Eur.J.Immunol., 54, 2024
|
|
6BX8
 
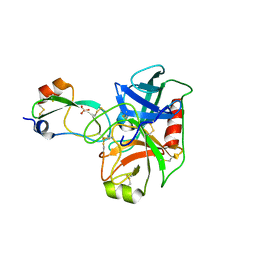 | | Human Mesotrypsin (PRSS3) Complexed with Tissue Factor Pathway Inhibitor Variant (TFPI1-KD1-K15R-I17C-I34C) | | Descriptor: | SULFATE ION, Tissue factor pathway inhibitor, Trypsin-3 | | Authors: | Coban, M, Sankaran, B, Cohen, I, Hockla, A, Papo, N, Radisky, E.S. | | Deposit date: | 2017-12-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J. Biol. Chem., 294, 2019
|
|
7QYH
 
 | |
7OR2
 
 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 4) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OSQ
 
 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OOU
 
 | | NaK C-DI mutant with Li+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OQ2
 
 | | NaK S-DI mutant soaked in Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
