3J7B
 
 | | Catalase solved at 3.2 Angstrom resolution by MicroED | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nannenga, B.L, Shi, D, Hattne, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2014-06-09 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Structure of catalase determined by MicroED.
Elife, 3, 2014
|
|
3K10
 
 | |
4C8L
 
 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the artificial base pair dNaM-d5SICS at the postinsertion site (sequence context 1) | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*AP*GP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8K
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a partially closed complex with the artificial base pair d5SICS-dNaMTP | | Descriptor: | ((2R,3S,5R)-3-hydroxy-5-(3-methoxynaphthalen-2-yl)methyl-tetrahydrogen-triphosphate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*AP*C*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C18
 
 | |
4CCH
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with d5SICS as templating nucleotide | | Descriptor: | 5'-D(*AP*AP*CP*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP* C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3', DNA POLYMERASE I, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8O
 
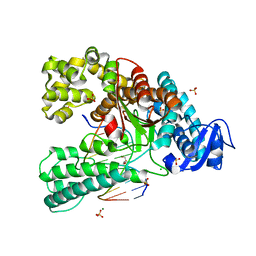 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*TP*TP*CP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*CP)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
3GQU
 
 | |
4C8N
 
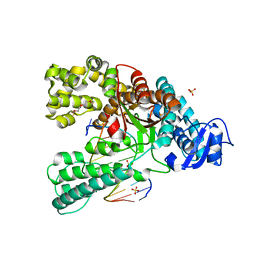 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 3) | | Descriptor: | LARGE FRAGMENT OF TAQ DNA POLYMERASE I, PRIMER, 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4BS2
 
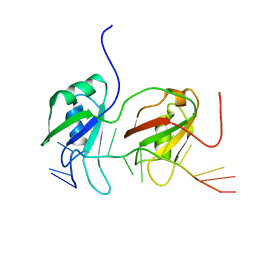 | | NMR structure of human TDP-43 tandem RRMs in complex with UG-rich RNA | | Descriptor: | 5'-R(*GP*UP*GP*UP*GP*AP*AP*UP*GP*AP*AP*UP)-3', TAR DNA-BINDING PROTEIN 43 | | Authors: | Lukavsky, P.J, Daujotyte, D, Tollervey, J.R, Ule, J, Stuani, C, Buratti, E, Baralle, F.E, Damberger, F.F, Allain, F.H.T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Nat.Struct.Mol.Biol., 20, 2013
|
|
1XHC
 
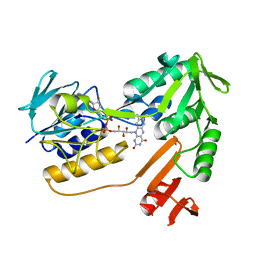 | | NADH oxidase /nitrite reductase from Pyrococcus furiosus Pfu-1140779-001 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase /nitrite reductase | | Authors: | Horanyi, P, Tempel, W, Weinberg, M.V, Liu, Z.-J, Shah, A, Chen, L, Lee, D, Sugar, F.J, Brereton, P.S, Izumi, M, Poole II, F.L, Shah, C, Jenney Jr, F.E, Arendall III, W.B, Rose, J.P, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-17 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | NADH oxidase /nitrite reductase from Pyrococcus furiosus Pfu-1140779-001
To be published
|
|
1HAU
 
 | | X-RAY STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE AT HIGH PH AND IN COPPER FREE FORM AT 1.9 A RESOLUTION | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Ellis, M.J, Dodd, F.E, Strange, R.W, Prudencio, M, Sawerseady, R.R, Hasnain, S.S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of a Blue Copper Nitrite Reductase at High Ph and in Copper-Free Form at 1.9 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HAW
 
 | | X-RAY STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE AT HIGH PH AND IN COPPER FREE FORM AT 1.9 A RESOLUTION | | Descriptor: | COPPER (I) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Ellis, M.J, Dodd, F.E, Strange, R.W, Prudencio, M, Sawerseady, R.R, Hasnain, S.S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of a Blue Copper Nitrite Reductase at High Ph and in Copper-Free Form at 1.9 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GS6
 
 | | Crystal structure of M144A mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, MAGNESIUM ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1GS8
 
 | | Crystal structure of mutant D92N Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1GS7
 
 | | Crystal structure of H254F mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1KBI
 
 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: Comparison with the Intact Wild-type Enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.-W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
1KBJ
 
 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: comparison with the Intact Wild-type Enzyme | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
1IU5
 
 | | X-ray Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IU6
 
 | | Neutron Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.6 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5K7R
 
 | | MicroED structure of trypsin at 1.7 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7Q
 
 | | MicroED structure of thaumatin at 2.5 A resolution | | Descriptor: | Thaumatin-1 | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7T
 
 | | MicroED structure of thermolysin at 2.5 A resolution | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7O
 
 | | MicroED structure of lysozyme at 1.8 A resolution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7N
 
 | | MicroED structure of tau VQIVYK peptide at 1.1 A resolution | | Descriptor: | VQIVYK | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
