7LPD
 
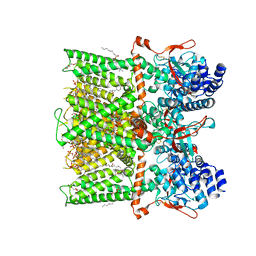 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 48 degrees Celsius, in an intermediate state, class 2 | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LPC
 
 | | Cryo-EM structure of full-length TRPV1 at 48 degrees Celsius | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, Phosphatidylinositol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7UWF
 
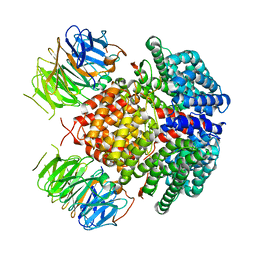 | | Human Rix1 sub-complex scaffold | | Descriptor: | Modulator of non-genomic activity of estrogen receptor, WD repeat-containing protein 18 | | Authors: | Gordon, J, Stanley, R.E. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM reveals the architecture of the PELP1-WDR18 molecular scaffold.
Nat Commun, 13, 2022
|
|
7UXA
 
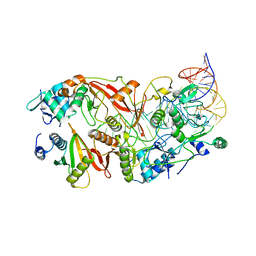 | | Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG | | Descriptor: | MAGNESIUM ION, RNA (78-MER), tRNA-splicing endonuclease subunit Sen15, ... | | Authors: | Stanley, R.E, Hayne, C.K. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3J70
 
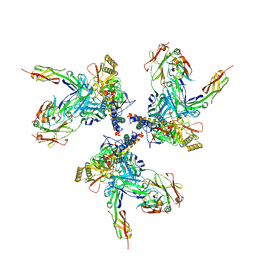 | | Model of gp120, including variable regions, in complex with CD4 and 17b | | Descriptor: | Envelope glycoprotein gp120, T-cell surface glycoprotein CD4, envelope glycoprotein gp41, ... | | Authors: | Rasheed, M, Bettadapura, R, Bajaj, C. | | Deposit date: | 2014-04-22 | | Release date: | 2015-08-26 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Computational Refinement and Validation Protocol for Proteins with Large Variable Regions Applied to Model HIV Env Spike in CD4 and 17b Bound State.
Structure, 23, 2015
|
|
7TU0
 
 | | Structure of the L. blandensis dGTPase bound to Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, dGTP triphosphohydrolase | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU1
 
 | | Structure of the L. blandensis dGTPase R37A mutant | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, dGTP triphosphohydrolase | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU2
 
 | | Structure of the L. blandensis dGTPase R37A mutant bound to Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, dGTP triphosphohydrolase | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU3
 
 | | Structure of the L. blandensis dGTPase del55-58 mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU4
 
 | | Structure of the L. blandensis dGTPase del55-58 mutant bound to Mn | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7K0R
 
 | | Nucleotide bound SARS-CoV-2 Nsp15 | | Descriptor: | PHOSPHATE ION, URIDINE-5'-MONOPHOSPHATE, Uridylate-specific endoribonuclease | | Authors: | Pillon, M.C, Stanley, R.E. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics.
Nat Commun, 12, 2021
|
|
7L06
 
 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L6M
 
 | |
7L02
 
 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L09
 
 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from masked 3D refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L6O
 
 | | Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 - gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2020-12-23 | | Release date: | 2021-04-14 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
8DTI
 
 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTG
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTH
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 2 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTF
 
 | | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
