2IZJ
 
 | | STREPTAVIDIN-BIOTIN PH 3.50 I4122 STRUCTURE | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZL
 
 | | STREPTAVIDIN-2-IMINOBIOTIN PH 7.3 I222 COMPLEX | | Descriptor: | 2-IMINOBIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZG
 
 | | STREPTAVIDIN-BIOTIN PH 2.0 I222 COMPLEX | | Descriptor: | BIOTIN, STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZH
 
 | | STREPTAVIDIN-BIOTIN PH 10.44 I222 COMPLEX | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
5D9L
 
 | | Rsk2 N-terminal Kinase in Complex with bis-phenol pyrazole | | Descriptor: | 4,4'-(1H-pyrazole-3,4-diyl)diphenol, GLYCEROL, Ribosomal protein S6 kinase alpha-3 | | Authors: | Appleton, B.A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Potent and Selective RSK Inhibitors as Biological Probes.
J.Med.Chem., 58, 2015
|
|
5D9K
 
 | | Rsk2 N-terminal Kinase in Complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, GLYCEROL, Ribosomal protein S6 kinase alpha-3 | | Authors: | Appleton, B.A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Potent and Selective RSK Inhibitors as Biological Probes.
J.Med.Chem., 58, 2015
|
|
6OJJ
 
 | |
6OP7
 
 | | Structure of oxidized VIM-20 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
6P5W
 
 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6PEI
 
 | | Structure of sorbitol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Sorbitol dehydrogenase (L-iditol 2-dehydrogenase) | | Authors: | Bailey-Elkin, B.A, Kohlmeier, M.G, Oresnik, I.J, Mark, B.L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the sorbitol dehydrogenase SmoS from Sinorhizobium meliloti 1021
Acta Crystallogr.,Sect.D, 77, 2021
|
|
6P5V
 
 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6PEJ
 
 | | Structure of sorbitol dehydrogenase from Sinorhizobium meliloti 1021 bound to sorbitol | | Descriptor: | Sorbitol dehydrogenase (L-iditol 2-dehydrogenase), sorbitol | | Authors: | Bailey-Elkin, B.A, Kohlmeier, M.G, Oresnik, I.J, Mark, B.L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the sorbitol dehydrogenase SmoS from Sinorhizobium meliloti 1021
Acta Crystallogr.,Sect.D, 77, 2021
|
|
6PSN
 
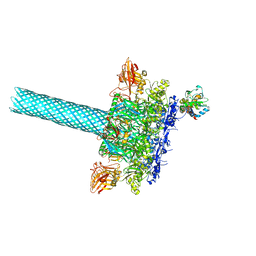 | | Anthrax toxin protective antigen channels bound to lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
6Q1M
 
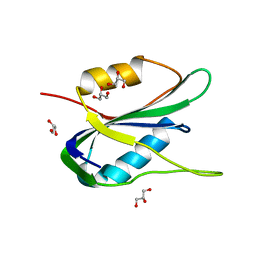 | | Crystal structure of the wheat dwarf virus Rep domain | | Descriptor: | GLYCEROL, Replication-associated protein | | Authors: | Litzau, L.A, Everett, B.A, Evans III, R.L, Shi, K, Tompkins, K, Gordon, W.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of the Wheat dwarf virus Rep domain.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6QQN
 
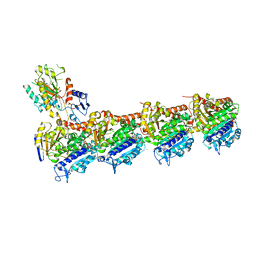 | | Tubulin-TH588 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Patterson, J.C, Joughin, B.A, Prota, A.E, Muehlethaler, T, Jonas, O.H, Whitman, M.A, Varmeh, S, Chen, S, Balk, S.P, Steinmetz, M.O, Lauffenburger, D.A, Yaffe, M.B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | VISAGE Reveals a Targetable Mitotic Spindle Vulnerability in Cancer Cells.
Cell Syst, 9, 2019
|
|
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
5J54
 
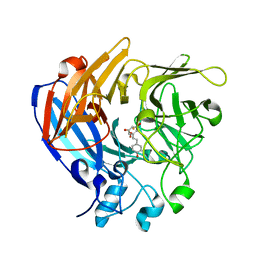 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5J55
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Carotenoid oxygenase, FE (III) ION, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5J53
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JG6
 
 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | Descriptor: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | Authors: | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0013 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
