3QVO
 
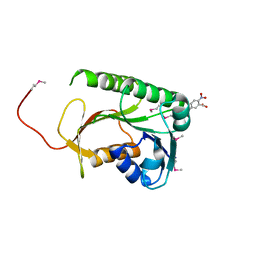 | | Structure of a Rossmann-fold NAD(P)-binding family protein from Shigella flexneri. | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, NmrA family protein | | Authors: | Cuff, M.E, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Rossmann-fold NAD(P)-binding family protein from Shigella flexneri.
TO BE PUBLISHED
|
|
3QSJ
 
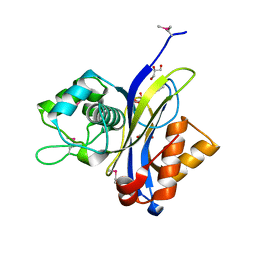 | | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius | | Descriptor: | CALCIUM ION, GLYCEROL, NUDIX hydrolase | | Authors: | Michalska, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius
To be Published
|
|
3QQZ
 
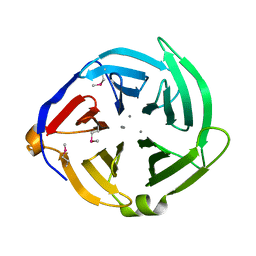 | | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein yjiK | | Authors: | Stein, A, Chhor, G, Nocek, B, Fenske, R.J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073
TO BE PUBLISHED
|
|
3NQR
 
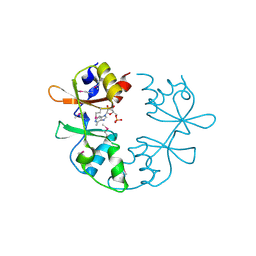 | | A putative CBS domain-containing protein from Salmonella typhimurium LT2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein corC | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Cui, H, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A putative CBS domain-containing protein from Salmonella typhimurium LT2.
To be Published
|
|
3OCM
 
 | | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative membrane protein, ... | | Authors: | Tan, K, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis
To be Published
|
|
3TEV
 
 | | The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 | | Descriptor: | Glycosyl hyrolase, family 3 | | Authors: | Chang, C, Hatzos-Skintges, C, Kohler, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-15 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1
To be Published
|
|
3QY3
 
 | | PA2801 protein, a putative Thioesterase from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Thioesterase | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the Pseudomonas aeruginosa hotdog-fold thioesterases PA5202 and PA2801.
Biochem.J., 444, 2012
|
|
3OOV
 
 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
3PAM
 
 | | Crystal structure of a domain of transmembrane protein of ABC-type oligopeptide transport system from Bartonella henselae str. Houston-1 | | Descriptor: | ETHANOL, Transmembrane protein | | Authors: | Nocek, B, Stein, A, Mack, J, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of a domain of transmembrane protein of ABC-type oligopeptide transport system from Bartonella henselae str. Houston-1"
TO BE PUBLISHED
|
|
3ONP
 
 | | Crystal Structure of tRNA/rRNA Methyltransferase SpoU from Rhodobacter sphaeroides | | Descriptor: | ACETIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-30 | | Release date: | 2010-09-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of tRNA/rRNA Methyltransferase SpoU from Rhodobacter sphaeroides
To be Published
|
|
3RCN
 
 | | Crystal Structure of Beta-N-Acetylhexosaminidase from Arthrobacter aurescens | | Descriptor: | ACETIC ACID, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Crystal Structure of Beta-N-Acetylhexosaminidase from Arthrobacter aurescens
To be Published
|
|
3QOM
 
 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum | | Descriptor: | 6-phospho-beta-glucosidase, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Michalska, K, Hatzos-Skintges, C, Bearden, J, Kohler, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3RNS
 
 | |
3RPZ
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with NADPH | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RHF
 
 | | Crystal structure of Polyphosphate Kinase 2 from Arthrobacter aurescens TC1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Feldmann, B, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Polyphosphate Kinase 2 from Arthrobacter aurescens TC1
TO BE PUBLISHED
|
|
3QOC
 
 | | Crystal structure of N-terminal domain (Creatinase/Prolidase like domain) of putative metallopeptidase from Corynebacterium diphtheriae | | Descriptor: | CHLORIDE ION, Putative metallopeptidase, SULFATE ION | | Authors: | Nocek, B, Stein, A, Marshall, N, Putagunta, R, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-09 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of N-terminal domain (Creatinase/Prolidase like domain) of putative metallopeptidase from Corynebacterium diphtheriae
TO BE PUBLISHED
|
|
3QTD
 
 | | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, PmbA protein | | Authors: | Tkaczuk, K.L, Chruszcz, M, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1
To be Published
|
|
3QUF
 
 | | The structure of a family 1 extracellular solute-binding protein from Bifidobacterium longum subsp. infantis | | Descriptor: | ACETIC ACID, Extracellular solute-binding protein, family 1, ... | | Authors: | Cuff, M.E, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-23 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a family 1 extracellular solute-binding protein from Bifidobacterium longum subsp. infantis
TO BE PUBLISHED
|
|
3RF6
 
 | | Crystal structure of glycerol-3 phosphate bound HAD-like phosphatase from Saccharomyces cerevisiae | | Descriptor: | CITRATE ANION, MAGNESIUM ION, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Nocek, B, Kuznetsova, K, Evdokimova, E, Savchenko, A, Iakunine, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-05 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of glycerol-3 phosphate bound HAD-like phosphatase from Saccharomyces cerevisiae
TO BE PUBLISHED
|
|
3RPD
 
 | | The structure of a B12-independent methionine synthase from Shewanella sp. W3-18-1 in complex with Selenomethionine. | | Descriptor: | GLYCEROL, Methionine synthase (B12-independent), SELENOMETHIONINE, ... | | Authors: | Cuff, M.E, Li, H, Hatzos-Skintges, C, Tesar, C, Bearden, J, Clancy, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a B12-independent methionine synthase from Shewanella sp. W3-18-1 in complex with Selenomethionine.
TO BE PUBLISHED
|
|
3RQ6
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQB
 
 | |
3RNL
 
 | | Crystal Structure of Sulfotransferase from Alicyclobacillus acidocaldarius | | Descriptor: | GLYCEROL, MAGNESIUM ION, Sulfotransferase | | Authors: | Kim, Y, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal Structure of Sulfotransferase from Alicyclobacillus acidocaldarius
To be Published
|
|
3RQ2
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with NADH | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQ1
 
 | | Crystal Structure of Aminotransferase Class I and II from Veillonella parvula | | Descriptor: | 2-OXOGLUTARIC ACID, Aminotransferase class I and II, CHLORIDE ION, ... | | Authors: | Kim, Y, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Aminotransferase Class I and II from Veillonella parvula
To be Published
|
|
