6KI7
 
 | | Pyrophosphatase mutant K30R from Acinetobacter baumannii | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Su, J. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
4KFV
 
 | |
5J9F
 
 | | Human GAR transformylase in complex with GAR and (4-{[2-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)ethyl]amino}benzoyl)-L-glutamic acid (AGF183) | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-(4-{[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)ethyl]amino}benzene-1-carbonyl)-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong, J, Deis, S.M, Dann III, C.E. | | Deposit date: | 2016-04-09 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tumor Targeting with Novel 6-Substituted Pyrrolo [2,3-d] Pyrimidine Antifolates with Heteroatom Bridge Substitutions via Cellular Uptake by Folate Receptor alpha and the Proton-Coupled Folate Transporter and Inhibition of de Novo Purine Nucleotide Biosynthesis.
J.Med.Chem., 59, 2016
|
|
6K21
 
 | | Pyrophosphatase from Acinetobacter baumannii | | Descriptor: | Inorganic pyrophosphatase, MAGNESIUM ION, SODIUM ION | | Authors: | Su, J. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
6K27
 
 | | Pyrophosphatase with PPi from Acinetobacter baumannii | | Descriptor: | DIPHOSPHATE, Inorganic pyrophosphatase, MAGNESIUM ION | | Authors: | Su, J. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
6KI8
 
 | | Pyrophosphatase mutant K149R from Acinetobacter baumannii | | Descriptor: | DIPHOSPHATE, Inorganic pyrophosphatase, MAGNESIUM ION | | Authors: | Su, J. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
6KJX
 
 | | Galectin-13 variant C138S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
6IH9
 
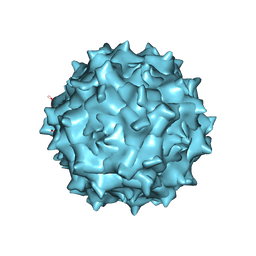 | | Adeno-Associated Virus 2 at 2.8 ang | | Descriptor: | Capsid protein VP1 | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
4LBF
 
 | |
3CQ0
 
 | | Crystal Structure of TAL2_YEAST | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative transaldolase YGR043C, ... | | Authors: | Huang, H, Niu, L, Teng, M. | | Deposit date: | 2008-04-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure and identification of NQM1/YGR043C, a transaldolase from Saccharomyces cerevisiae
Proteins, 73, 2008
|
|
4LB1
 
 | |
8HKH
 
 | |
7BRS
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA02 | | Descriptor: | 8-[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-indol-1-ium-1-yl]octanoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-03-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6M0A
 
 | | The heme-bound structure of the chloroplast protein At3g03890 | | Descriptor: | AT3G03890 protein, AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Liu, L. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Arabidopsis locus AT3G03890 encodes a dimeric beta-barrel protein implicated in heme degradation.
Biochem.J., 2020
|
|
6M09
 
 | |
9HCF
 
 | | Mouse mitoribosome large subunit assembly intermediate bound to NSUN4, METRF4, GTPBP7, GTPBP10 and the MALSU-L0R8F8-mtACP complex with uL16m, State B2 (SAMC knock-out) | | Descriptor: | 16S rRNA (1432-MER), 5-cytosine rRNA methyltransferase NSUN4, Acyl carrier protein, ... | | Authors: | Singh, V, Rorbach, J, Freyer, C, Amunts, A, Wredenberg, A. | | Deposit date: | 2024-11-08 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | The mitochondrial methylation potential gates mitoribosome assembly.
Nat Commun, 16, 2025
|
|
7SFR
 
 | | Unmethylated Mtb Ribosome 50S with SEQ-9 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Xing, Z, Cui, Z, Zhang, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2021-10-04 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
4XS3
 
 | | Crystal structure of a metabolic reductase with (E)-1-benzyl-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one | | Descriptor: | (E)-1-benzyl-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, B, Wu, F, Jiang, H, Kogiso, M, Yao, Y, Zhou, C, Li, X, Song, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.291 Å) | | Cite: | Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds.
J.Med.Chem., 58, 2015
|
|
9IK9
 
 | | Cryo-EM Structure of SST analogs bond SSTR1-Gi complex | | Descriptor: | (4J2)(DCY)(DTY)(DTR)K(DVA)(DCY)(ALO)(NH2), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Du, Y. | | Deposit date: | 2024-06-26 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into the binding modes of lanreotide and pasireotide with somatostatin receptor 1.
Acta Pharm Sin B, 15, 2025
|
|
4XRX
 
 | | Crystal structure of a metabolic reductase with (E)-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one | | Descriptor: | 5-[(E)-(1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl]pyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, B, Wu, F, Jiang, H, Kogiso, M, Yao, Y, Zhou, C, Li, X, Song, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds.
J.Med.Chem., 58, 2015
|
|
9IK8
 
 | | Cryo-EM Structure of SSTR1-Gi SST analogs complex | | Descriptor: | DTR-LYS-TY5-PHA-A1D5E-004, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Du, Y. | | Deposit date: | 2024-06-26 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural insights into the binding modes of lanreotide and pasireotide with somatostatin receptor 1.
Acta Pharm Sin B, 15, 2025
|
|
8W4P
 
 | | Structure of PSII-FCPII-I/J/K complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
8W4O
 
 | | Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
7E6Q
 
 | | Crystal structure of influenza A virus neuraminidase N5 complexed with 4'-phenyl-1,2,3-triazolylated oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-acetamido-3-pentan-3-yloxy-5-(4-phenyl-1,2,3-triazol-1-yl)cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, P.F, Babayemi, O.O, Li, C.N, Fu, L.F, Zhang, S.S, Qi, J.X, Lv, X, Li, X.B. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of 5'-substituted 1,2,3-triazolylated oseltamivir derivatives as potent influenza neuraminidase inhibitors.
Rsc Adv, 11, 2021
|
|
1E9K
 
 | | The structure of the RACK1 interaction sites located within the unique N-terminal region of the cAMP-specific phosphodiesterase, PDE4D5. | | Descriptor: | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Bolger, G.B, Smith, K.J, McCahill, A, Hyde, E.I, Steele, M.R, Houslay, M.D. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H NMR structural and functional characterisation of a cAMP-specific phosphodiesterase-4D5 (PDE4D5) N-terminal region peptide that disrupts PDE4D5 interaction with the signalling scaffold proteins, beta-arrestin and RACK1.
Cell. Signal., 19, 2007
|
|
