7VYJ
 
 | | The structure of GdmN in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYO
 
 | | The structure of GdmN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZN
 
 | | The structure of GdmN in complex with carbamoyl adenylate intermediate and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZU
 
 | | The structure of GdmN Y82F mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GdmN, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZY
 
 | | The structure of GdmN complex with AMP and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
5EK7
 
 | |
8IA9
 
 | |
8IAA
 
 | |
8HVR
 
 | |
7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPB
 
 | | SARS-CoV-2 Omicron Variant RBD complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPC
 
 | | The second RBD of SARS-CoV-2 Omicron Variant in complexed with RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPD
 
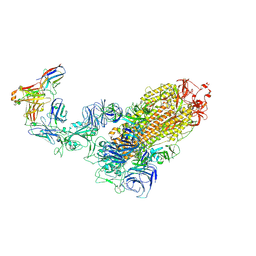 | | SARS-CoV-2 Omicron Variant S Trimer complexed with one JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPF
 
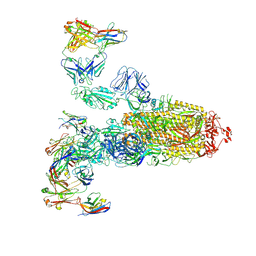 | | SARS-CoV-2 Omicron Variant S Trimer complexed with three JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPE
 
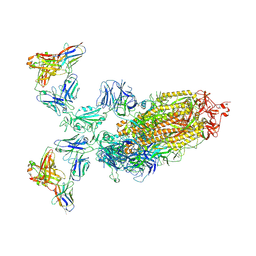 | | SARS-CoV-2 Omicron Variant S Trimer complexed with two JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WRV
 
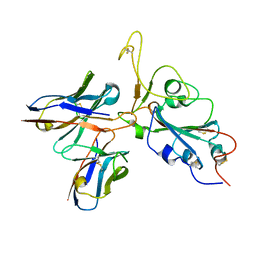 | | The interface of JMB2002 Fab binds to SARS-CoV-2 Omicron Variant S | | Descriptor: | JMB2002 Fab heavy chain, JMB2002 Fab light chain, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
6IYO
 
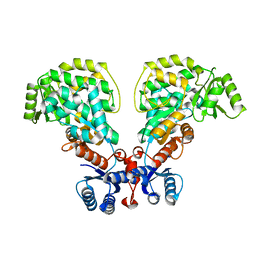 | |
6J0U
 
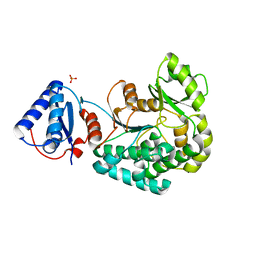 | |
6IYT
 
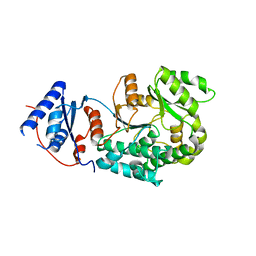 | |
6IYR
 
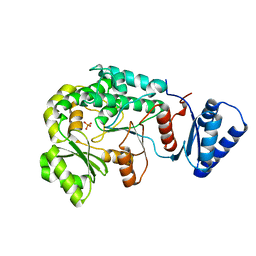 | |
3Q9H
 
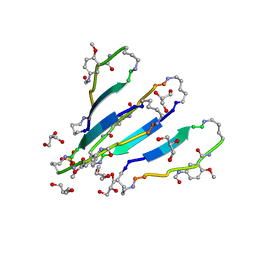 | | LVFFA segment from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | 1,4-BUTANEDIOL, Cyclic pseudo-peptide LVFFA(ORN)(HAO)LK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
6IIH
 
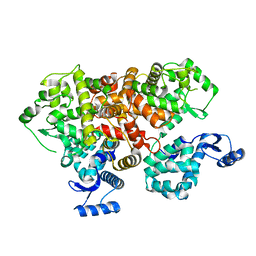 | | crystal structure of mitochondrial calcium uptake 2(MICU2) | | Descriptor: | CALCIUM ION, Endolysin,Calcium uptake protein 2, mitochondrial | | Authors: | Shen, Q, Wu, W, Zheng, J, Jia, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | The crystal structure of MICU2 provides insight into Ca2+binding and MICU1-MICU2 heterodimer formation.
Embo Rep., 20, 2019
|
|
3Q9G
 
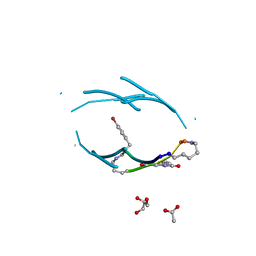 | | VQIVY segment from Alzheimer's tau displayed on 42-membered macrocycle scaffold | | Descriptor: | ACETIC ACID, Cyclic pseudo-peptide VQIV(4BF)(ORN)(HAO)KL(ORN), GLYCEROL | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9J
 
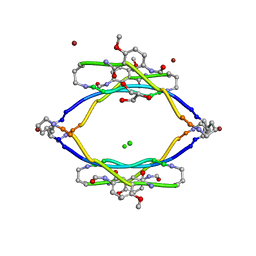 | | AIIFL segment derived from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | CHLORIDE ION, Cyclic pseudo-peptide AIIFL(ORN)(HAO)YK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
