5KF4
 
 | |
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6T5V
 
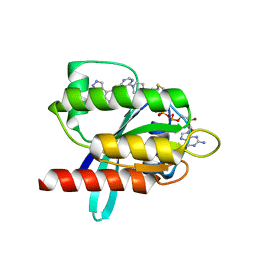 | | KRasG12C ligand complex | | Descriptor: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
7N2O
 
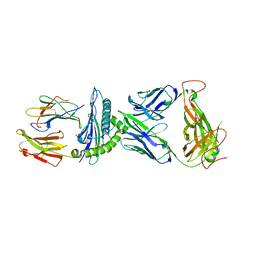 | | AS4.2-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2Q
 
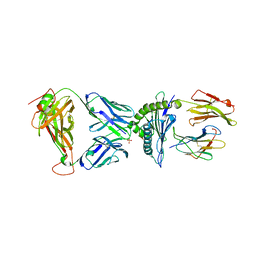 | | AS4.3-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2S
 
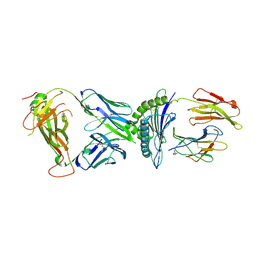 | | AS3.1-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2R
 
 | | AS4.3-PRPF3-HLA*B27 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2N
 
 | | TCR-antigen complex AS4.2-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2P
 
 | | AS4.3-RNASEH2b-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
3PL9
 
 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | Deposit date: | 2010-11-14 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8HPU
 
 | |
8HQ7
 
 | |
8HPF
 
 | |
8HPV
 
 | |
8HP9
 
 | |
8HPQ
 
 | |
1U6Q
 
 | | Substituted 2-Naphthamadine inhibitors of Urokinase | | Descriptor: | TRANS-6-(2-PHENYLCYCLOPROPYL)-NAPHTHALENE-2-CARBOXAMIDINE, Urokinase-type plasminogen activator | | Authors: | Bruncko, M, McClellan, W, Wendt, M.D, Sauer, D.R, Geyer, A, Dalton, C.R, Kaminski, M.K, Nienaber, V.L, Rockway, T.R, Giranda, V.L. | | Deposit date: | 2004-07-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Naphthamidine urokinase plasminogen activator inhibitors with improved pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4RTH
 
 | | The crystal structure of PsbP from Zea mays | | Descriptor: | Membrane-extrinsic protein of photosystem II PsbP | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
4RTI
 
 | | The crystal structure of PsbP from Spinacia oleracea | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Oxygen-evolving enhancer protein 2, ... | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
6W5S
 
 | | NPC1 structure in GDN micelles at pH 8.0 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5V
 
 | | NPC1-NPC2 complex structure at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5R
 
 | | NPC1 structure in Nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5U
 
 | | NPC1 structure in GDN micelles at pH 5.5, conformation b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5T
 
 | | NPC1 structure in GDN micelles at pH 5.5, conformation a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
3RJX
 
 | | Crystal Structure of Hyperthermophilic Endo-Beta-1,4-glucanase | | Descriptor: | Endoglucanase FnCel5A | | Authors: | Zheng, B.S, Yang, W, Zhao, X.Y, Wang, Y.G, Lou, Z.Y, Rao, Z.H, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of hyperthermophilic endo-beta-1,4-glucanase: implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
