3SVD
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: bromide complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
3TAP
 
 | |
3TAQ
 
 | |
3SST
 
 | |
3SVE
 
 | |
3SRY
 
 | |
3SVB
 
 | |
3SSK
 
 | |
3SS0
 
 | |
3SSH
 
 | |
3TAN
 
 | |
3THV
 
 | |
3TI0
 
 | |
6A2W
 
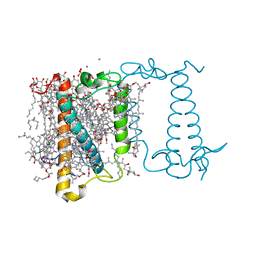 | | Crystal structure of fucoxanthin chlorophyll a/c complex from Phaeodactylum tricornutum | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W, Yu, L.J, Kuang, T.Y, Shen, J.R. | | Deposit date: | 2018-06-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for blue-green light harvesting and energy dissipation in diatoms.
Science, 363, 2019
|
|
6J0Z
 
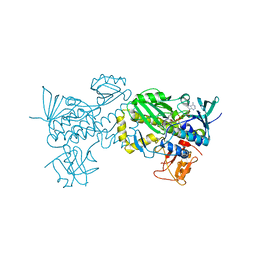 | | Crystal structure of AlpK | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative angucycline-like polyketide oxygenase | | Authors: | Wang, W, Liu, Y, Liang, H. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Crystal structure of AlpK: An essential monooxygenase involved in the biosynthesis of kinamycin
Biochem. Biophys. Res. Commun., 510, 2019
|
|
7VP8
 
 | | Crystal structure of ferritin from Ureaplasma urealyticum | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin-like diiron domain-containing protein | | Authors: | Wang, W, Liu, X, Wang, Y, Fu, D, Wang, H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Distinct structural characteristics define a new subfamily of Mycoplasma ferritin
Chin.Chem.Lett., 33, 2022
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | Descriptor: | CHLORIDE ION, SODIUM ION, Vps75 | | Authors: | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
4JGV
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
3SOV
 
 | | The structure of a beta propeller domain in complex with peptide S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low-density lipoprotein receptor-related protein 6, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3SOB
 
 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a FAB | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 6, antibody heavy chain, ... | | Authors: | Wang, W, Bourhis, E, Tam, C, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3SOQ
 
 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a DKK1 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dickkopf-related protein 1, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
2VG4
 
 | | Rv2361 native | | Descriptor: | UNDECAPRENYL PYROPHOSPHATE SYNTHETASE | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VG1
 
 | | Rv1086 E,E-farnesyl diphosphate complex | | Descriptor: | FARNESYL DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
4XZC
 
 | | The crystal structure of Kupe virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
2VG0
 
 | | Rv1086 citronellyl pyrophosphate complex | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, SHORT-CHAIN Z-ISOPRENYL DIPHOSPHATE SYNTHETASE | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
