2BR1
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | 2-[5,6-BIS-(4-METHOXY-PHENYL)-FURO[2,3-D]PYRIMIDIN-4-YLAMINO]-ETHANOL, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-04-29 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRM
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | 3-AMINO-3-BENZYL-[4.3.0]BICYCLO-1,6-DIAZANONAN-2-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRN
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | (2R)-1-[(5,6-DIPHENYL-7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)AMINO]PROPAN-2-OL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRH
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | N-(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)GLYCINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRB
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | 2-[(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)AMINO]ETHANOL, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2C3K
 
 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | 4-[3-(1H-BENZIMIDAZOL-2-YL)-1H-INDAZOL-6-YL]-2-METHOXYPHENOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
2C3J
 
 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | DEBROMOHYMENIALDISINE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFITE ION | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
2VP8
 
 | | Structure of Mycobacterium tuberculosis Rv1207 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPTEROATE SYNTHASE 2 | | Authors: | Gengenbacher, M, Xu, T, Niyomwattanakit, P, Spraggon, G, Dick, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Biochemical and Structural Characterization of the Putative Dihydropteroate Synthase Ortholog Rv1207 of Mycobacterium Tuberculosis.
Fems Microbiol.Lett., 287, 2008
|
|
1Z8S
 
 | | DnaB binding domain of DnaG (P16) from Bacillus stearothermophilus (residues 452-597) | | Descriptor: | DNA primase | | Authors: | Syson, K, Thirlway, J, Hounslow, A.M, Soultanas, P, Waltho, J.P. | | Deposit date: | 2005-03-31 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the helicase-interaction domain of the primase DnaG: a model for helicase activation
Structure, 13, 2005
|
|
4AXK
 
 | | CRYSTAL STRUCTURE OF subHisA from the thermophile Corynebacterium efficiens | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-((5'-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO)IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, GLYCEROL | | Authors: | Noda-Garcia, L, Camacho-Zarco, A.R, Medina-Ruiz, S, Verduzco-Castro, E.A, Gaytan, P, Carrillo-Tripp, M, Fulop, V, Barona-Gomez, F. | | Deposit date: | 2012-06-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of Substrate Specificity in a Recipient'S Enzyme Following Horizontal Gene Transfer.
Mol.Biol.Evol., 30, 2013
|
|
3ZUQ
 
 | | Crystal structure of an engineered botulinum neurotoxin type B - derivative, LC-B-GS-Hn-B | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, ZINC ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3ZUR
 
 | | Crystal structure of an engineered botulinum neurotoxin type A- SNARE23 derivative, LC0-A-SNAP25-Hn-A | | Descriptor: | BOTULINUM NEUROTOXIN TYPE A, SYNAPTOSOMAL-ASSOCIATED PROTEIN, SULFATE ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3ZH9
 
 | | Bacillus subtilis DNA clamp loader delta protein (YqeN) | | Descriptor: | DELTA, GLYCEROL, SULFATE ION | | Authors: | Suwannachart, C, Sedelnikova, S, Soultanas, P, Oldham, N.J, Rafferty, J.B. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structure and Assembly of the Bacillus Subtilis Clamp-Loader Complex and its Interaction with the Replicative Helicase.
Nucleic Acids Res., 41, 2013
|
|
3ZUS
 
 | | Crystal structure of an engineered botulinum neurotoxin type A- SNARE23 derivative, LC-A-SNAP23-Hn-A | | Descriptor: | BOTULINUM NEUROTOXIN TYPE A, SYNAPTOSOMAL-ASSOCIATED PROTEIN 23, ZINC ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2Y1W
 
 | | CRYSTAL STRUCTURE OF COACTIVATOR ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1) IN COMPLEX WITH SINEFUNGIN AND INDOLE INHIBITOR | | Descriptor: | 2-{4-[3-FLUORO-2-(2-METHOXYPHENYL)-1H-INDOL-5-YL] PIPERIDIN-1-YL}-N-METHYLETHANAMINE, HISTONE-ARGININE METHYLTRANSFERASE CARM1, SINEFUNGIN | | Authors: | Sack, J.S, Thieffine, S, Bandiera, T, Fasolini, M, Duke, G.J, Jayaraman, L, Kish, K.F, Klei, H.E, Purandare, A.V, Rosettani, P, Troiani, S, Xie, D, Bertrand, J.A. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Carm1 Inhibition by Indole and Pyrazole Inhibitors
Biochem.J., 436, 2011
|
|
2Y1X
 
 | | CRYSTAL STRUCTURE OF COACTIVATOR ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1) IN COMPLEX WITH SINEFUNGIN AND INDOLE INHIBITOR | | Descriptor: | CHLORIDE ION, HISTONE-ARGININE METHYLTRANSFERASE CARM1, N-(3-{5-[5-(1H-INDOL-4-YL)-1,3,4-OXADIAZOL-2-YL]-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL}BENZYL)-L-ALANINAMIDE, ... | | Authors: | Sack, J.S, Thieffine, S, Bandiera, T, Fasolini, M, Duke, G.J, Jayaraman, L, Kish, K.F, Klei, H.E, Purandare, A.V, Rosettani, P, Troiani, S, Xie, D, Bertrand, J.A. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Carm1 Inhibition by Indole and Pyrazole Inhibitors
Biochem.J., 436, 2011
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
7XIM
 
 | |
7XJF
 
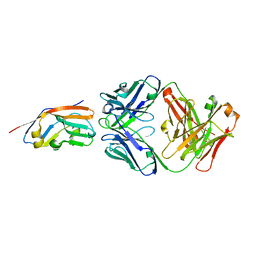 | | Crystal structure of 6MW3211 Fab in complex with CD47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2022-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Blockade of dual immune checkpoint inhibitory signals with a CD47/PD-L1 bispecific antibody for cancer treatment.
Theranostics, 13, 2023
|
|
6TVH
 
 | |
6TX2
 
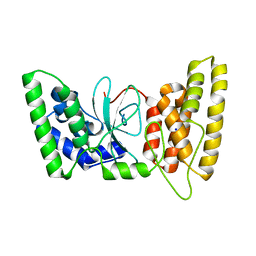 | | Human HPF1 | | Descriptor: | Histone PARylation factor 1, SODIUM ION | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
9XIM
 
 | |
8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8AHG
 
 | |
8AHH
 
 | |
